

















When InDels with certain characteristics, described below, are present in the guidance track provided to the Local Realignment tool, false positive insertions and deletions may be introduced into read mappings.
This issue can arise where, for a given guidance InDel, the sequence of the insertion or deletion at that position is similar to the adjacent reference sequence (figure 1). Reads that support the reference sequence may then be re-aligned to the guidance InDel instead of the reference. Using the resulting read mapping for variant detection may then lead to these regions being detected as variants with higher read support than the guidance variant had.
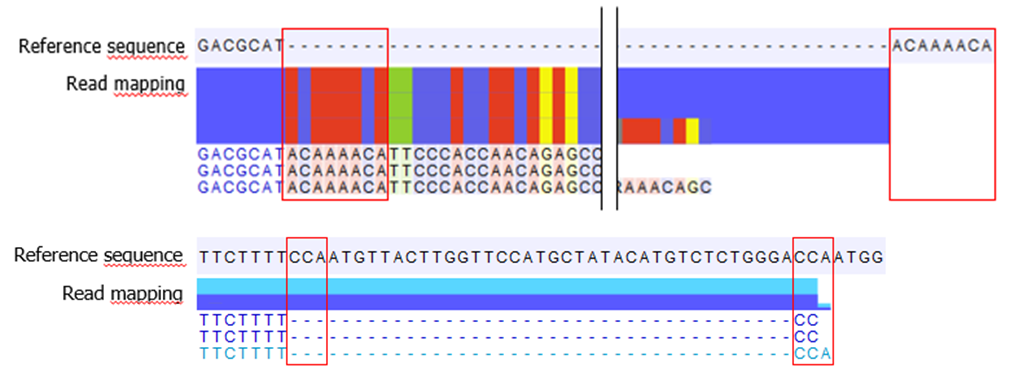
Figure 1. Examples of read mappings where Local Realignment introduced a false-positive insertion (top) or deletion (bottom). The sequence at the beginning of each is similar to the reference sequence immediately after the InDel (red boxes).
This issue has been observed in data from QIAseq SARS-CoV-2 panels, as well as in data from other QIAseq panels, analyzed using template workflows provided by the Biomedical Genomics Analysis plugin when run on affected software versions (see below).
We recommend upgrading to a version of the software not affected by this issue.
If continuing to use affected software versions, the risk that the issue will occur can be reduced by including only high-confidence variants in a guidance track for Local Realignment.
When using affected software versions, the risk that the issue will occur can be reduced by including only high-confidence variants in a guidance track for Local Realignment.
For example, in workflows designed to detect variants at germline frequencies, remove InDels with a frequency of 5% or lower, by using the Filter on Custom Criteria tool, and provide this track for guidance to Local Realignment (figure 2).
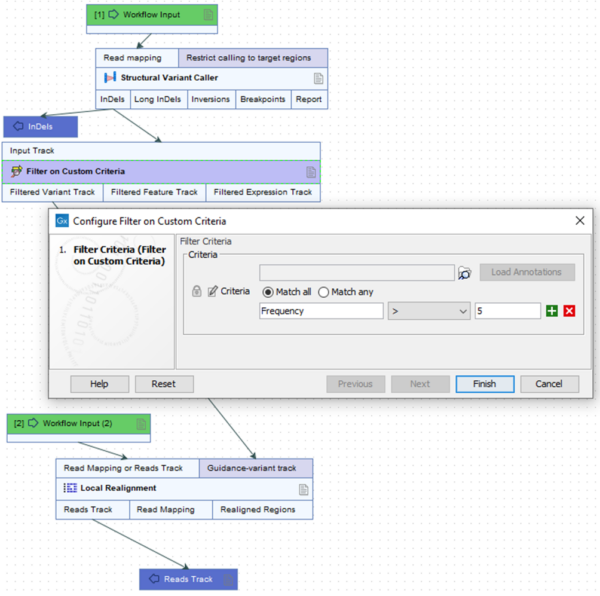
Figure 2. Removing low frequency InDels from a track intended for use as a guidance can decrease the risk of observing the issue described.
• CLC Genomics Workbench and CLC Genomics Server 22.0 and 22.0.1
Local Realignment is included in all QIAGEN template workflows for the identification of variants in DNA sequencing data.
This issue was addressed in CLC Genomics Workbench 22.0.2 and CLC Genomics Server 22.0.2.