Latest improvements for QIAGEN OmicSoft Suite
OmicSoft Lands API v1.7 and MyQDI release update
We are happy to announce that QIAGEN OmicSoft Lands API v1.7 is now available for download, along with the latest updates to the OmicSoft Lands databases.
Transfer Assist for AWS
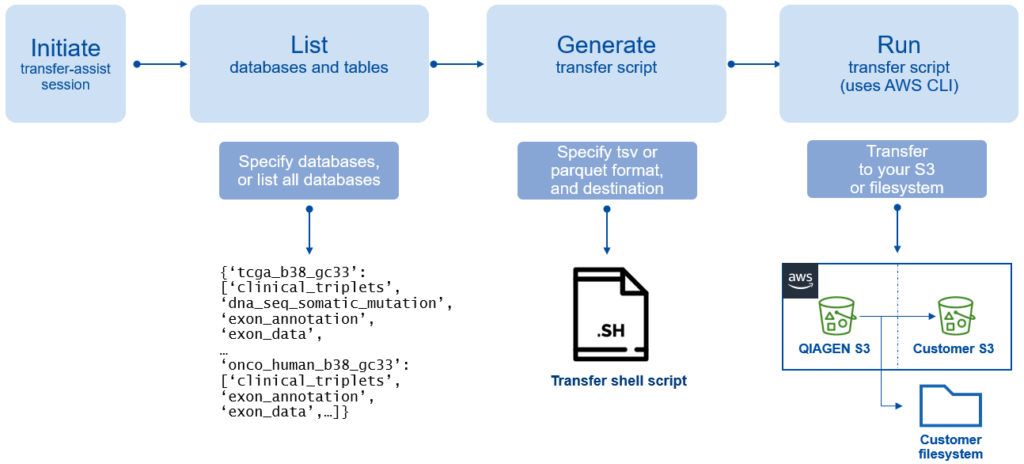
Figure 1. Diagram of “Transfer Assist for AWS”.
We released the utility “Transfer Assist for AWS”, which allows you to use the AWS CLI to quickly transfer full Lands databases from our S3 buckets directly to user buckets or drive locations. You no longer must download them to temporary locations, such as EBS volumes, only to upload them back to an S3 location.
Download the Transfer Assist for AWS utility, QIAGEN OmicSoft Lands API v1.7 and the latest user manual at https://my.qiagendigitalinsights.com .
New datasets available
A major update to LINCS is now available as LINCS_B38_GC33, with over 160,000 statistical comparisons ready to use to explore gene signatures of genetic perturbations. LINCS_B38_GC33 replaces the legacy LINCS_B37 database.
Learn more about LINCS Land here.
In addition, the latest Lands updates to OncoHuman, HumanDisease and MouseDisease are available for querying in OmicSoft Lands API and download. Learn more about the latest updates here.
OmicSoft Suite v12.6 Release Notes
Top Improvements:
· Curated gene signatures
· Dynamic correlation methods
· Expanded support for Amazon Web Services (AWS) analysis
· New data types supported in Land Explorer visualizations
· QIAGEN OmicSoft Lands API update
QIAGEN OmicSoft Suite v12.6 is now available, with several new improvements to support your NGS data analysis and exploration of curated ‘omics data. Check out the full details in the OmicSoft Help menu’s “Change Log”. Important note: If you aren’t prompted to update and given a preview of latest improvements, you may not be using the latest OmicSoft Studio Updater.
If you connect to a dedicated OmicSoft Server instance, please remind your administrator to update your OmicSoft Server. The next update to TCGA Land will include methylation data, but requires OmicSoft Suite v12.4+, and this latest release brings even more new capabilities.
Now would be a good time for your OmicSoft Server administrator to review your ArrayServer.cfg file for the latest recommended configuration parameters, including updated computer images (“AMIs”) for cloud-based NGS data analysis, support for faster downloads of FASTQ files from SRA, and user authorization options.
Search Lands data with curated gene signatures from QIAGEN Knowledge Base
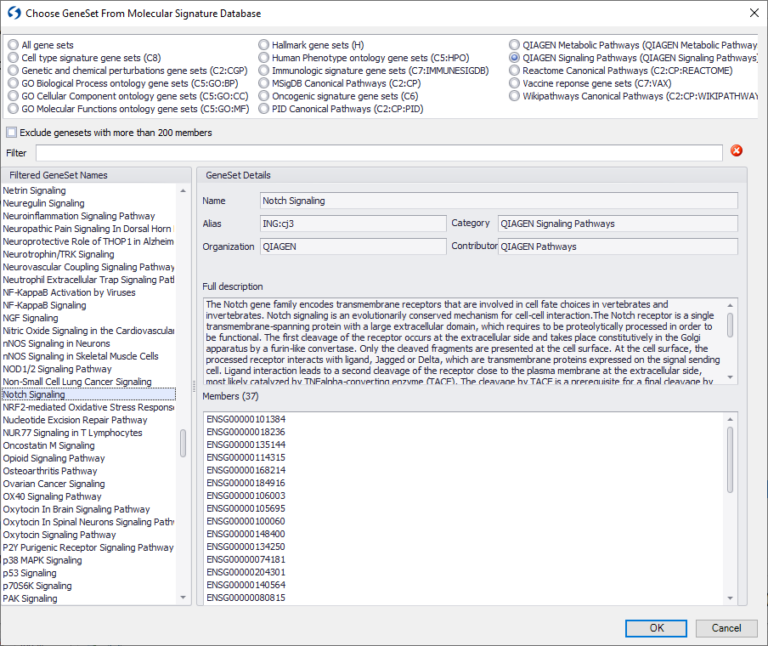
You can now search across QIAGEN OmicSoft Lands datasets using the Human Gencode.v33, Mouse Gencode.v17 or Mouse Gencode.v24 models using thousands of gene signatures corresponding to curated signaling and metabolic pathways from QIAGEN Knowledge Base, new gene signatures from Reactome and the Broad Institute Molecular Signatures Database (MSigDB).
Figure 1. Example of choosing gene signatures from QIAGEN Knowledge Base.
Find top correlated protein expression
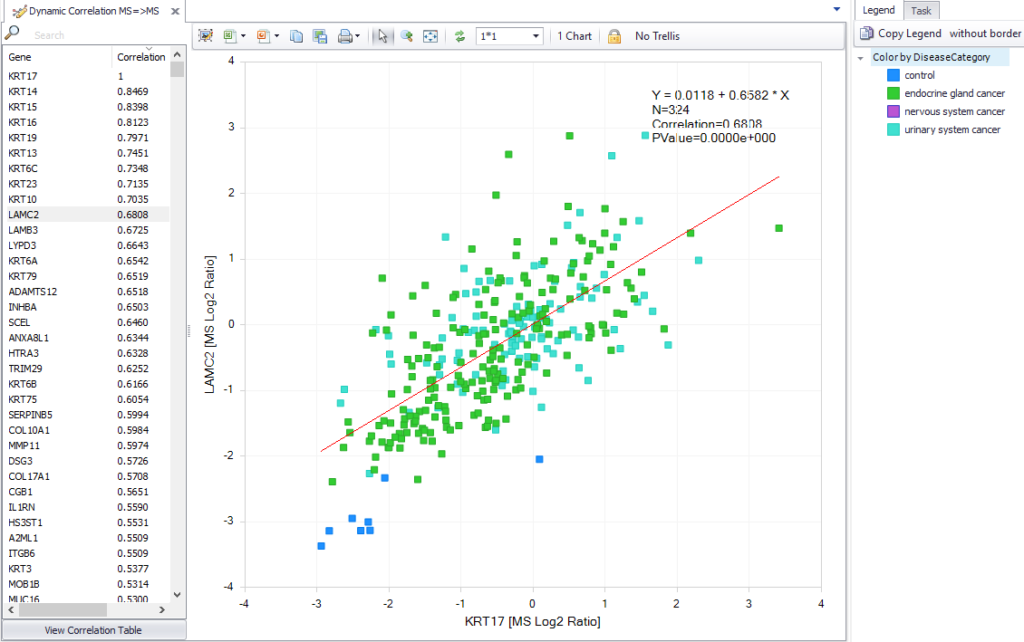
We’ve improved one of the most powerful capabilities in QIAGEN OmicSoft Lands to allow you to find correlated protein expression. You can find these data in ClinicalProteomicTumor, TCGA, GTEx, BeatAML and CCLE databases.
Figure 2. Improved identification of correlated protein expression.
Find co-expressed genes in curated single-cell data
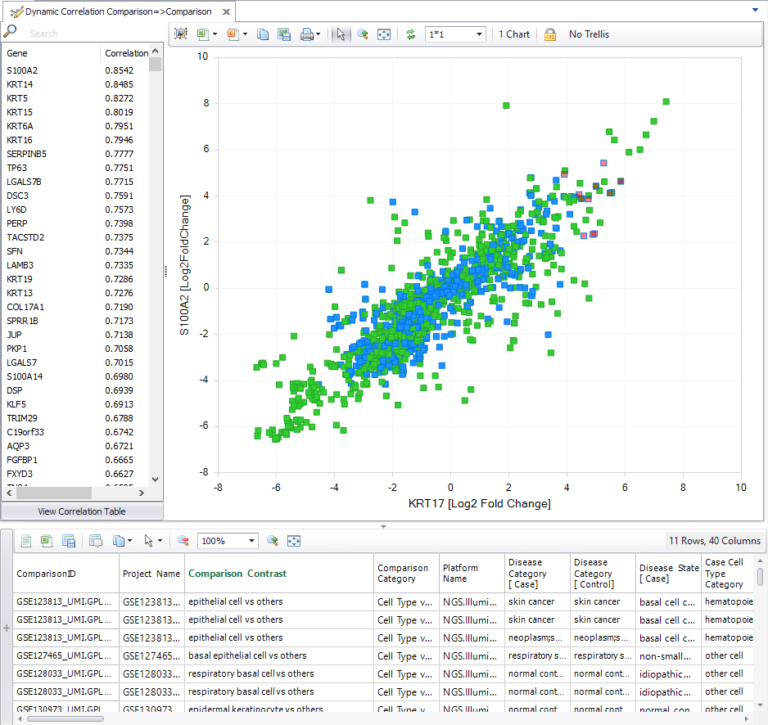
You can now look for co-regulated genes across statistical comparisons, including single-cell comparisons. OmicSoft Single Cell Land datasets are manually curated for specific cell types from each cell cluster, and differential expression is calculated within analyses for cluster-enriched and cell type-enriched genes. You can now use the dynamic comparison correlation tool to find other genes that are up- and down-regulated in the same clusters and cell types as your gene of interest.
Figure 3. Visualization of the dynamic comparison correlation tool with single cell comparisons.
Land “Show Summary Information” shows fold-change between two sample groups
In QIAGEN OmicSoft Land visualizations with exactly two groups, “Show Summary Information” will calculate the log2 Fold Change between the median of each group. This tool provides another metric to assess target differences when viewing look-up tables of gene expression between two sample groups, such as two differnt tissues, disease vs. control or treatment vs. control.
Figure 4. Visualization of the show summary information tool.
Nitro AMI support for Ubuntu20
Cloud NGS jobs are now supported on M5-class AWS instances built on Ubuntu20 AMIs, in addition to AL2 AMIs. Be sure to update your ArrayServer.cfg file with preferred instance types, using OAlignInstanceType and OSummaryInstanceType.
Other improvements you won’t want to miss
OmicSoft Lands API update
OmicSoft Lands API v1.6 is now available from https://my.qiagendigitalinsights.com . Use this latest version and the available demo notebooks to follow along with webinars at https://tv.qiagenbioinformatics.com.
Support for upcoming LINCS Land database updates
In anticipation of a major update to LINCS, including thousands more comparisons and using the latest genome/gene model, OmicSoft Suite v12.6 includes several performance improvements. Be sure that your OmicSoft Administrator updates your server if you plan to explore LINCS data.
Support for Methylation data in TCGA Land database
The winter release of OmicSoft OncoLand includes the integration of methylation array data, which requires OmicSoft Suite v12.4+.
New visualizations in Land Explorer web UI – Support MS and Methylation Views
Land Explorer’s latest version now supports sample-level MS and Methylation Array views.
Latest content available in QIAGEN OmicSoft Lands
Recent updates to OmicSoft Lands Human Disease, Mouse Disease, OncoHuman and ATCC Cell Line Human/Mouse are available in OmicSoft Studio, Land Explorer and OmicSoft Lands API. Learn more in the content update release notes.
Get in touch with us
Please reach out to ts-bioinformatics@qiagen.com for any technical questions or issues or if you are interested in a training.
OmicSoft Suite v12.5 Release Notes
OmicSoft Suite v12.5 is now available, with several new improvements for NGS analysis and exploration of curated omics data. Check out the full details in the OmicSoft Help menu’s “Change Log”.
With the new FASTQ download function, you can now simultaneously retrieve associated metadata from both the SRA and GEO repositories, ready to attach to your project files. The new module also supports previewing the data to ensure you chose the right project.
- Preview GEO metadata for samples
- Simultaneous download of GEO and SRA metadata, ready to append to analyzed samples
- Easily append metadata to OmicData and NgsData objects
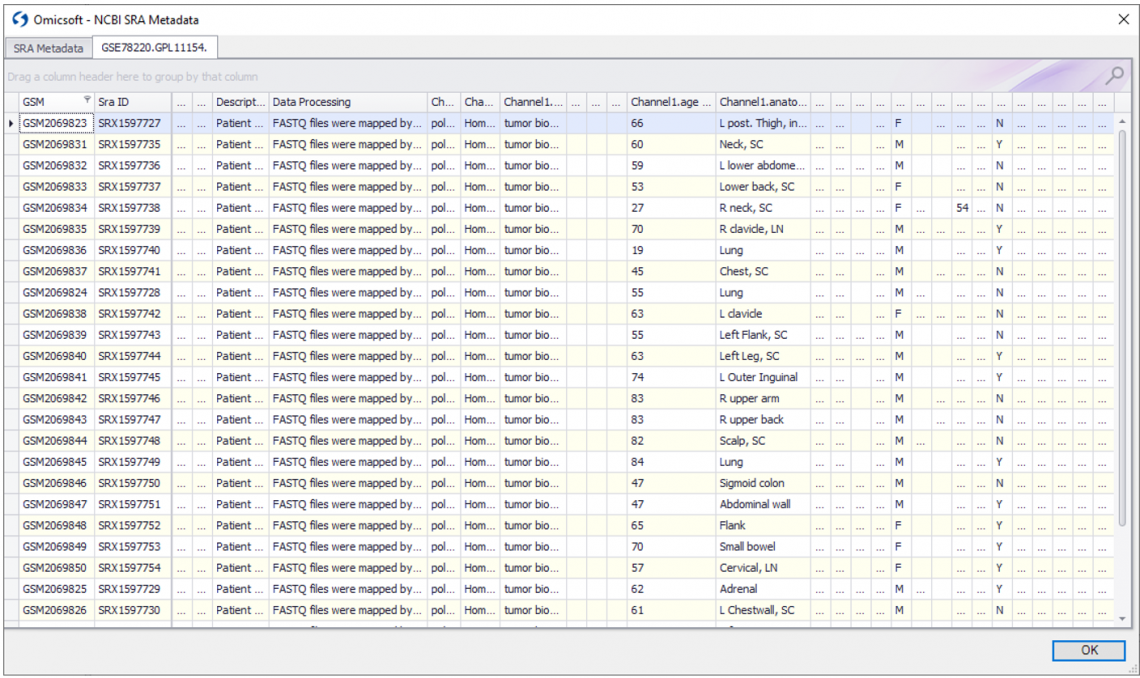
Figure 1. Preview both SRA and GEO metadata for your SRA downloads before downloading, to make sure that you are downloading the correct datasets.
Mass Spectrometry proteomics comparisons
The new Land database “ClinicalProteomicTumor” incorporates multi-omics cancer profiling studies from the CPTAC repository. These data focus on Mass Spectrometry (MS) data, but also includes RNA-seq, miRNA-seq, mutation, CNV, and methylation data.
In addition to sample-level protein expression data, you can explore statistical comparisons at the protein level, and compare differential expression patterns between protein and RNA between cohorts.
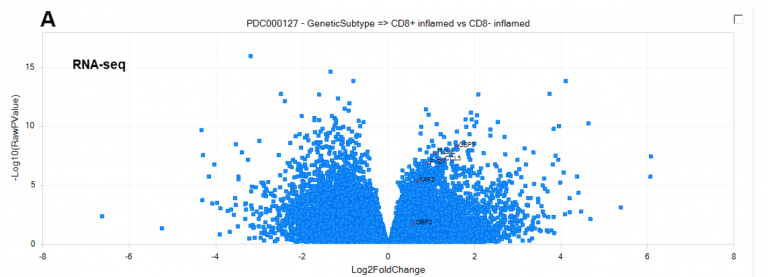
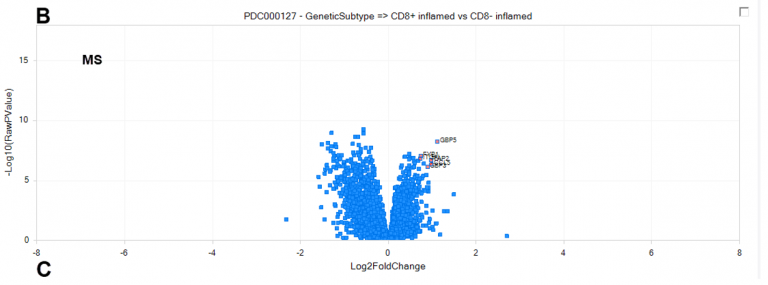
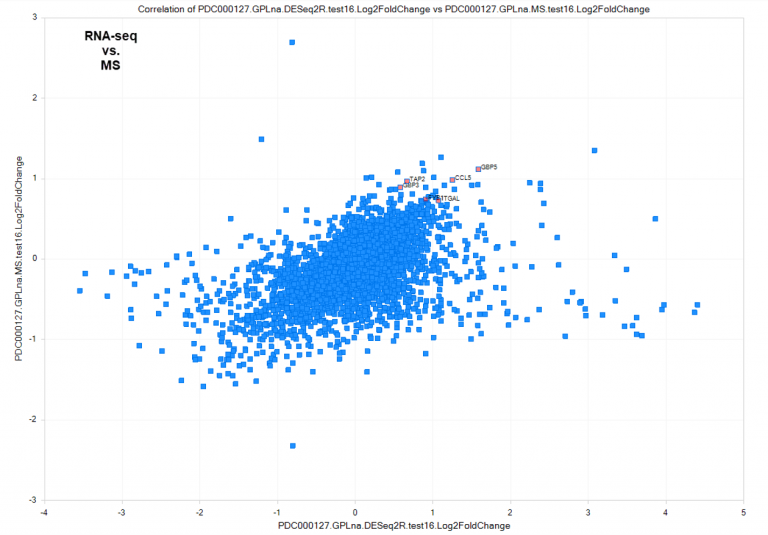
Figure 2. New MS comparisons allow deeper interrogation of proteomics datasets curated by the Omicsoft team, such as CD8+ inflamed (interferon-γ signaling) vs CD8− inflamed (platelet degranulation) tumors in clear cell Renal Cell Carcinoma (ccRCC) from PDC000127, at the RNA and protein level. (A) Differentially-regulated genes at the RNA level, calculated by DEseq2. (B) Differentially-regulated genes at the protein level, calculated by general linear model. Top up-regulated proteins were selected to illustrate concordance with RNA-level up-regulation. (C) Comparison of direction and magnitude between RNA (X-axis) and Protein (Y-axis) differential expression. Genes measured only at the RNA-level are plotted with a fold-change=0 on the Y-axis.
OmicSoft Studio installer supports improved options for updating to new versions of the software.
- Preview the key improvements- before installing
- Update, delay, or turn off updates
- Administrators can deploy static non-updating versions to scientists
OmicSoft Lands API update
The latest update to OmicSoft Lands API (1.3) now supports REST interface to submit SQL queries. In addition, several improvements to query submission performance and bug fixes were addressed. Download the latest client and release notes details from my.qiagendigitalinsights.com.
Please reach out to ts-bioinformatics@qiagen.com for any technical questions or issues, or if you are interested in a training!
OmicSoft Suite v12.4 Release Notes
OmicSoft Suite v12.4 delivers important bug fixes and improvements requested by customers.
Key improvements include:
- Software installation management
- New MSI installer allowing application administrators to install static versions in App Stores: https://resources.omicsoft.com/software/OmicSoftStudioInstaller/OmicSoft_Studio_Installer_12.4.1.1.msi
- New logo for OmicSoft Studio
- OmicSoft Server on the Cloud
- Improved download of FASTQ from the NCBI SRA repository (server on the Cloud configuration required)
- Parallel download of FASTQ files from NCBI to your S3 bucket, server, or local client
- Download SRA metadata simultaneously with your data
- Multiple AWS instance types can be specified in oscript
- For advanced users of oscript and Server on the Cloud, /InstanceType supports comma-delimited lists of instance types
- OmicSoft Server on the Cloud — IAM Instance Profile authentication supported
- Alternative to including Access Key in ArrayServer.cfg configuration
- OmicSoft Server on the Cloud AMIs are no longer specified by default. Please select the appropriate AMI and specify Ami and AmiSnapshot parameters in the [Cloud] section of your ArrayServer.cfg
- Ubuntu 20 AMIs
- Amazon Linux 2 AMIs
- Server management
- LDAP integration now supports restriction to specific user groups
- SIEM logging of server activity supported – contact your account manager for details
- Data resources
- Single Cell Land improved download speed
- 1 annotator (includes 1000Genomes and ExAc) is now available for annotating VCFs on Human Genome 38
- Additional improvements and bug fixes, enumerated in the Change Log (visible under the OmicSoft Studio Help menu > View Change Log
Compatibility
OmicSoft Suite v12.4 components are compatible with previous OmicSoft Suite v12 components.
- OmicSoft Server v12.4 is compatible with OmicSoft Studio v12.3, v12.2, and v12.1
- OmicSoft Studio v12.4 is compatible with OmicSoft Server v12.3, v12.2, and v12.1
- oshell v12.4 is compatible with OmicSoft Server v12.3, v12.2, and v12.1
- LandExplorer v.4.3 is compatible with OmicSoft Server v12.3, v12.2, and v12.1
OmicSoft Suite v12 components are not compatible with OmicSoft Suite v11.7 or older components.
Retired Functionalities
- The BAS Server was retired starting OmicSoft Suite v12.0.
- OmicSoft Viewer was retired starting OmicSoft Suite v12.0, and link-sharing functionalities are removed.
- The legacy “Studio on the Cloud” feature has been deprecated for Studio-only users. However, OmicSoft Server supports Cloud NGS analysis with the Server on the Cloud Contact your account manager if you are interested in learning more.
Known issues
- Users cannot create mapped folder pointing to /users/ virtual subfolders
- If a Cloud-mapped S3 bucket contains an object '/', it will be displayed as a folder '/' that cannot be opened. Delete this folder in the S3 bucket.
- Users cannot search for transcripts names when creating omic data queries
- If updating from OmicSoft Server v12.1 or v12.2 on Windows, previous installation services must be uninstalled before the new WiX installer for OmicSoft Server Service 12.4 is installed.
- The new WiX installer for v12.4 should be used for new installations of OmicSoft Server Service or directly updating from OmicSoft Server v12.3.
- Update Cloud Folder Mapping will create a new folder instead of updating the name of the existing cloud folder.
- On Mac OmicSoft Studio, some charts cannot be exported using Export to PowerPoint or Export to Excel
- After filtering and recoloring cohorts in Survival views, colors are sometimes inverted between legend and X-axis labels.
- RNA-seq→TMA view is visible in HPA_B37 although no samples are available with RNA-seq + TMA data; no data are displayed.
- Web site for ArrayExpress is not rendered properly in Add Expression Data > ArrayExpress Experiment
- Open in PowerPoint always opens a new PowerPoint® file; this function no longer supports appending to an open file.
If you have further questions, please contact your local QIAGEN® representative or contact our Technical Support Center at www.qiagen.com/support/technical-support
OmicSoft Suite v12.3 Release Notes
QIAGEN OmicSoft Lands API expands access to curated ‘omics data
The new QIAGEN OmicSoft Lands API enables powerful searches across QIAGEN OmicSoft Lands data. Query data on any metadata field across Land databases, without needing to ingest data into your own SQL database.

Figure 1. QIAGEN OmicSoft Lands API schematic. Queries are submitted using SQL syntax via Python or R packages, which are processed by the QIAGEN OmicSoft Lands API query engine to find matching data within the QIAGEN curated ‘omics Land databases. Matching data are returned to the Python/R environment for analysis and visualization. Retrieved data can be downloaded as flat files or directly loaded into Python/R data frames.
Data
The QIAGEN OmicSoft Lands API provides direct access to OmicSoft’s collections of deeply-curated datasets, taking advantage of extensive controlled vocabularies and our human-centered curation approach to unlock powerful cross-project queries.
Learn more: Why is a curated ‘omics data portal important?
Data structure
OmicSoft’s structured tables of metadata and data can be queried on one or more fields and joined to return exactly the data needed to answer your biomarker-related questions.
QIAGEN OmicSoft OncoLand and DiseaseLand datasets have structured and curated metadata at project, sample and comparison level and are linked to ‘omics measurements for each sample, such as RNA-seq gene expression (gene, transcript, exon, junction, fusion and mutation), microarray expression (probe and gene), miRNA-seq, DNA-seq mutation calls, copy numbers and more.

Figure 2. Schema of QIAGEN OmicSoft OncoLand and DiseaseLand datasets. Curated metadata (left) are represented within multiple tables with keys to join at the project and sample level, as well as comparisons between sample groups. Each curated data type (middle) is represented by a separate table, which can be filtered on associated metadata, data values or associated annotation (right).
Single-cell datasets within QIAGEN OmicSoft Single Cell Land databases include structured and curated metadata at project, sample, cluster, cell and comparison level, linking to gene expression (raw and normalized counts) for each cell, along with cell coordinates from pre-computed dimension reduction analyses.

Figure 3. Schema of single-cell RNA-seq datasets in QIAGEN OmicSoft Single Cell Land databases. In addition to curated information at the project, sample and comparison level (left), single-cell datasets include metadata and data describing cells and cell clusters (right).
Access to curated data
The initial release provides access to subscribed datasets within QIAGEN OmicSoft DiseaseLand, OncoLand and Single Cell Land databases, including the following:
- 650,000+ manually curated samples
- 1400+ diseases and 700+ tissues represented across 9000+ ‘omics projects
- Rich harmonized metadata described across 6000+ metadata fields, allowing powerful queries to capture relevant samples across studies
- 124,000+ comparisons from custom statistical models to reveal differentially expressed gene signatures across studies
API client system requirements
We offer simple-to-install clients supporting Python (Python version 3.7 or higher) and R (R version 3.6.0 or higher). These clients are used to submit SQL queries to retrieve data slices from the hosted database.
Ask your Sales Representative to add the OmicSoft Lands API to your license.