Latest improvements for QIAGEN OmicSoft Suite
OmicSoft Lands API v1.7 and MyQDI release update
We are happy to announce that QIAGEN OmicSoft Lands API v1.7 is now available for download, along with the latest updates to the OmicSoft Lands databases.
Transfer Assist for AWS
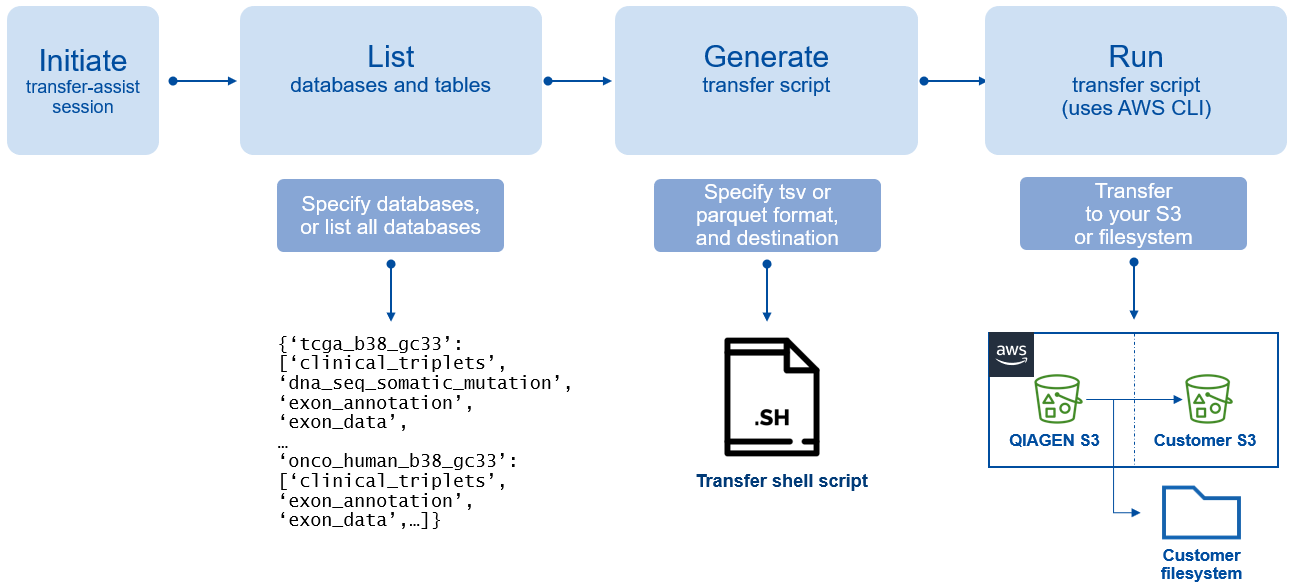
Figure 1. Diagram of “Transfer Assist for AWS”.
We released the utility “Transfer Assist for AWS”, which allows you to use the AWS CLI to quickly transfer full Lands databases from our S3 buckets directly to user buckets or drive locations. You no longer must download them to temporary locations, such as EBS volumes, only to upload them back to an S3 location.
Download the Transfer Assist for AWS utility, QIAGEN OmicSoft Lands API v1.7 and the latest user manual at https://my.qiagendigitalinsights.com .
New datasets available
A major update to LINCS is now available as LINCS_B38_GC33, with over 160,000 statistical comparisons ready to use to explore gene signatures of genetic perturbations. LINCS_B38_GC33 replaces the legacy LINCS_B37 database.
Learn more about LINCS Land here.
In addition, the latest Lands updates to OncoHuman, HumanDisease and MouseDisease are available for querying in OmicSoft Lands API and download. Learn more about the latest updates here.
OmicSoft Suite v12.6 Release Notes
Top Improvements:
· Curated gene signatures
· Dynamic correlation methods
· Expanded support for Amazon Web Services (AWS) analysis
· New data types supported in Land Explorer visualizations
· QIAGEN OmicSoft Lands API update
QIAGEN OmicSoft Suite v12.6 is now available, with several new improvements to support your NGS data analysis and exploration of curated ‘omics data. Check out the full details in the OmicSoft Help menu’s “Change Log”. Important note: If you aren’t prompted to update and given a preview of latest improvements, you may not be using the latest OmicSoft Studio Updater.
If you connect to a dedicated OmicSoft Server instance, please remind your administrator to update your OmicSoft Server. The next update to TCGA Land will include methylation data, but requires OmicSoft Suite v12.4+, and this latest release brings even more new capabilities.
Now would be a good time for your OmicSoft Server administrator to review your ArrayServer.cfg file for the latest recommended configuration parameters, including updated computer images (“AMIs”) for cloud-based NGS data analysis, support for faster downloads of FASTQ files from SRA, and user authorization options.
Search Lands data with curated gene signatures from QIAGEN Knowledge Base
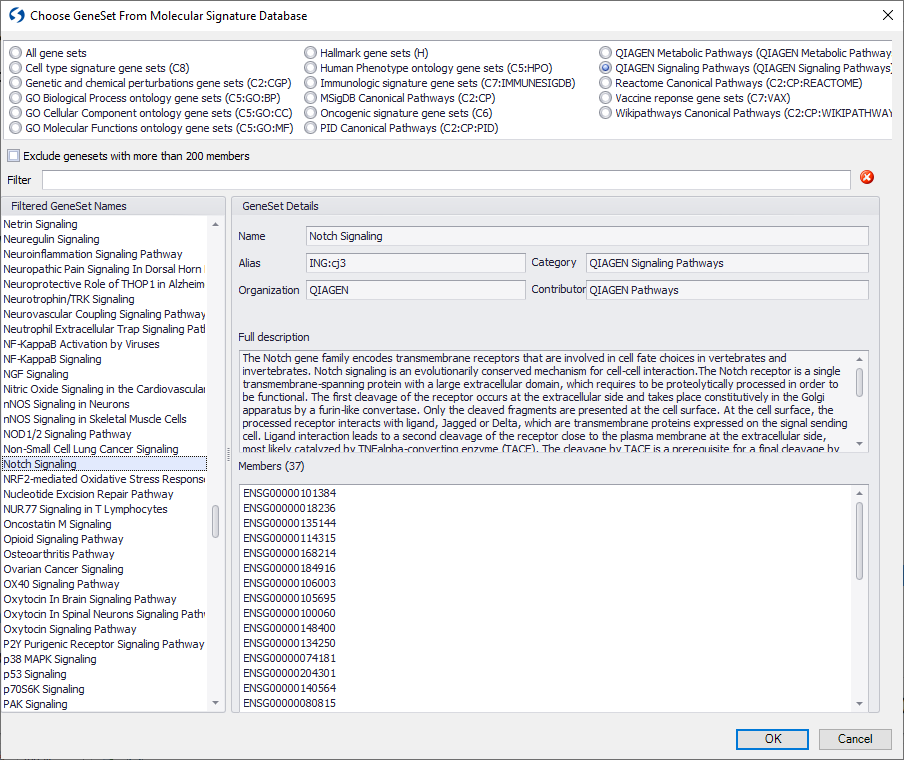
You can now search across QIAGEN OmicSoft Lands datasets using the Human Gencode.v33, Mouse Gencode.v17 or Mouse Gencode.v24 models using thousands of gene signatures corresponding to curated signaling and metabolic pathways from QIAGEN Knowledge Base, new gene signatures from Reactome and the Broad Institute Molecular Signatures Database (MSigDB).
Figure 1. Example of choosing gene signatures from QIAGEN Knowledge Base.
Find top correlated protein expression
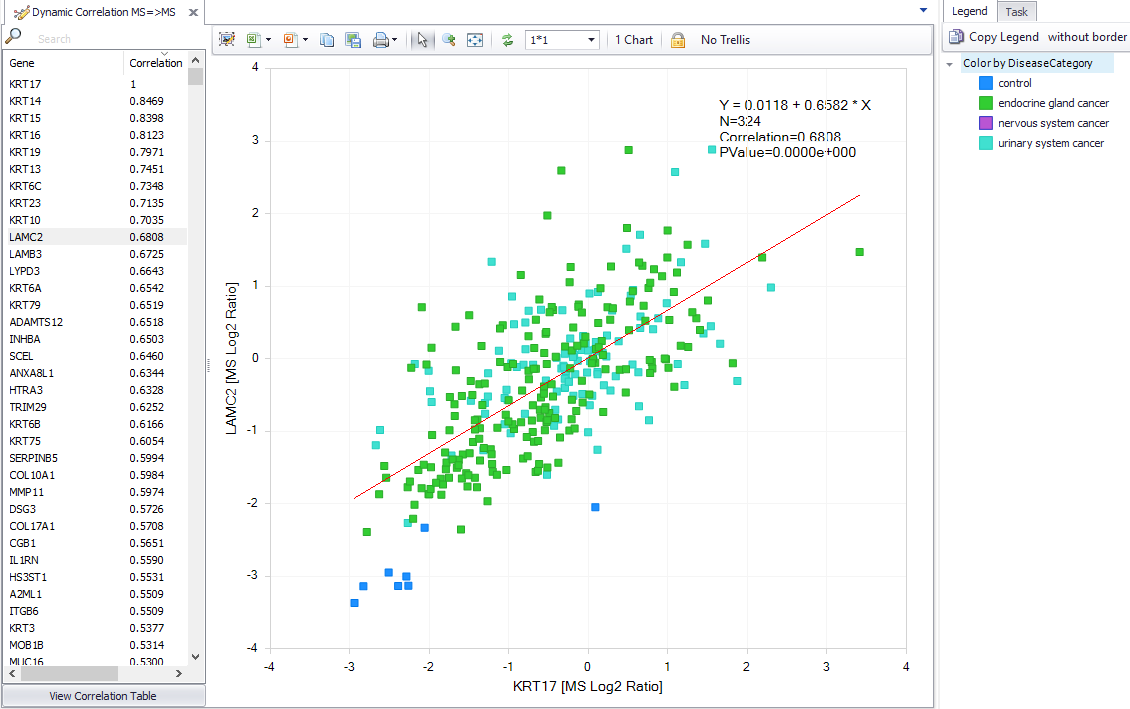
We’ve improved one of the most powerful capabilities in QIAGEN OmicSoft Lands to allow you to find correlated protein expression. You can find these data in ClinicalProteomicTumor, TCGA, GTEx, BeatAML and CCLE databases.
Figure 2. Improved identification of correlated protein expression.
Find co-expressed genes in curated single-cell data
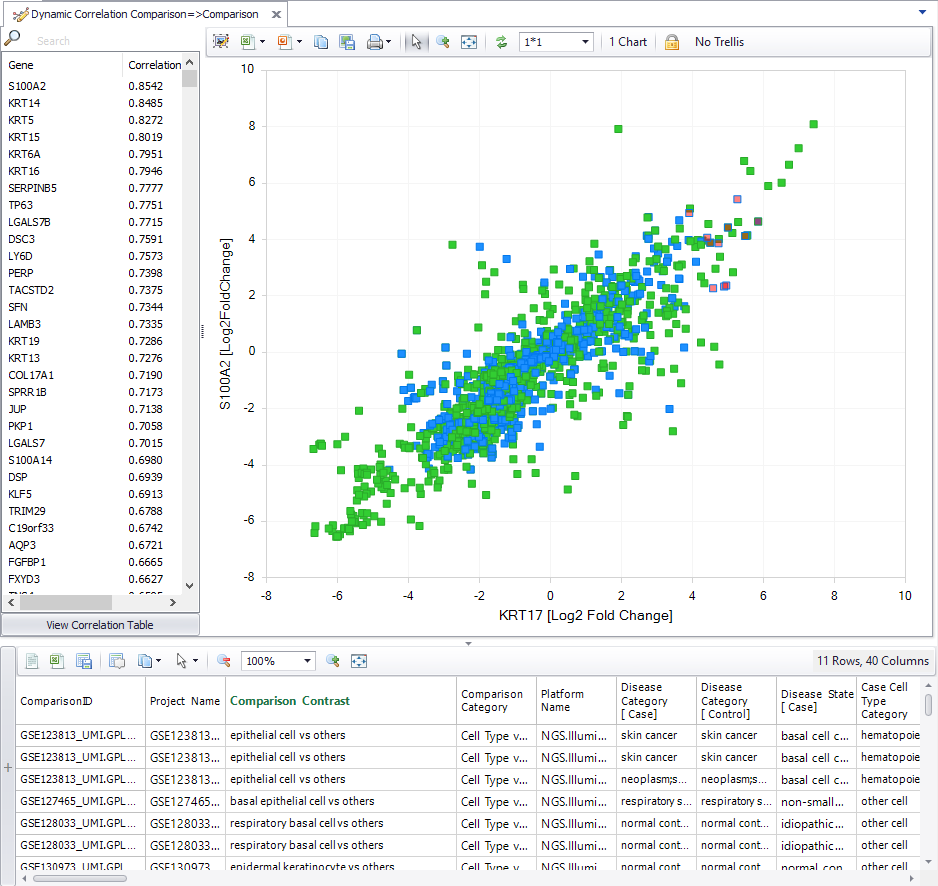
You can now look for co-regulated genes across statistical comparisons, including single-cell comparisons. OmicSoft Single Cell Land datasets are manually curated for specific cell types from each cell cluster, and differential expression is calculated within analyses for cluster-enriched and cell type-enriched genes. You can now use the dynamic comparison correlation tool to find other genes that are up- and down-regulated in the same clusters and cell types as your gene of interest.
Figure 3. Visualization of the dynamic comparison correlation tool with single cell comparisons.
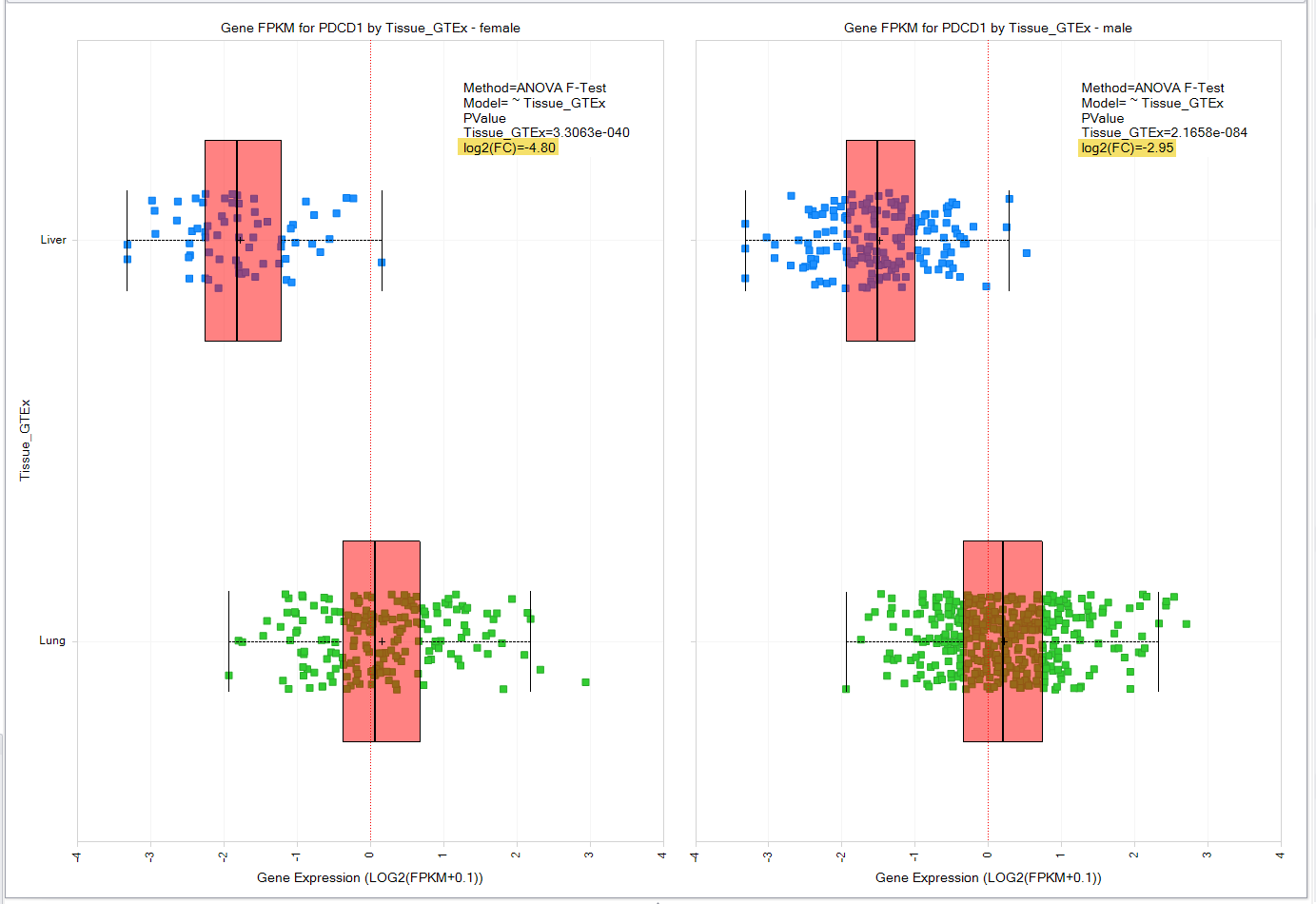
Land “Show Summary Information” shows fold-change between two sample groups
In QIAGEN OmicSoft Land visualizations with exactly two groups, “Show Summary Information” will calculate the log2 Fold Change between the median of each group. This tool provides another metric to assess target differences when viewing look-up tables of gene expression between two sample groups, such as two differnt tissues, disease vs. control or treatment vs. control.
Figure 4. Visualization of the show summary information tool.
Nitro AMI support for Ubuntu20
Cloud NGS jobs are now supported on M5-class AWS instances built on Ubuntu20 AMIs, in addition to AL2 AMIs. Be sure to update your ArrayServer.cfg file with preferred instance types, using OAlignInstanceType and OSummaryInstanceType.
Other improvements you won’t want to miss
OmicSoft Lands API update
OmicSoft Lands API v1.6 is now available from https://my.qiagendigitalinsights.com . Use this latest version and the available demo notebooks to follow along with webinars at https://tv.qiagenbioinformatics.com.
Support for upcoming LINCS Land database updates
In anticipation of a major update to LINCS, including thousands more comparisons and using the latest genome/gene model, OmicSoft Suite v12.6 includes several performance improvements. Be sure that your OmicSoft Administrator updates your server if you plan to explore LINCS data.
Support for Methylation data in TCGA Land database
The winter release of OmicSoft OncoLand includes the integration of methylation array data, which requires OmicSoft Suite v12.4+.
New visualizations in Land Explorer web UI – Support MS and Methylation Views
Land Explorer’s latest version now supports sample-level MS and Methylation Array views.
Latest content available in QIAGEN OmicSoft Lands
Recent updates to OmicSoft Lands Human Disease, Mouse Disease, OncoHuman and ATCC Cell Line Human/Mouse are available in OmicSoft Studio, Land Explorer and OmicSoft Lands API. Learn more in the content update release notes.
Get in touch with us
Please reach out to ts-bioinformatics@qiagen.com for any technical questions or issues or if you are interested in a training.

 Archive
Archive