NGS Secondary and Tertiary Analysis
Deliver clear and confident precision oncology reports
QCI Interpret for Oncology is the industry’s only automated FASTQ to final report solution for precision oncology NGS testing powered by AI-enabled literature searches and state-of-the-art manual curation
SOLID TUMORS | HEMATOLOGY ONCOLOGY | LIQUID BIOPSY | HEREDITARY CANCER
QCI Interpret for Oncology
An end-to-end solution for NGS variant analysis, interpretation and reporting
QCI Interpret for Oncology is an end-to-end solution for NGS data analysis, interpretation, and reporting that helps clinical diagnostic labs scale the process of FASTQ to final report. Comprised of two integrated software applications with an addtional in-software option to send rare or novel variants to QIAGEN’s on-demand variant interpreation service, QCI Interpret for Oncology alleviates the complexities of regulating in-house bioinformatics pipelines to reduce turnaround time, simplify variant interpretation, and support confident decisions.
QCI Interpret for Oncology
Cloud-based NGS secondary analysis that is an optional service for labs needing a FASTQ to VCF solution.
Clinical decision support software (tertiary analysis) with on-demamd variant interpretation services for labs needing a VCF to final report solution.
Automate FASTQ to final report
1. Upload raw FASTQ sequencing files
• Select pre-configured analysis pipelines in QCI Secondary Analysis (optional service for FASTQ to VCF)
• Generate high-quality variant calls, visualize results, and send VCF files directly to QCI Interpret

2. Upload VCFs and enter patient data
• Select workflow in QCI Interpret
• Upload variant files for a patient sample and enter relevant case details, such as patient diagnosis

3. View list of auto-classified variants
- View list of computed AMP/ASCO/CAP classifications for each variant
- Filter and rank by variants by significance
- View supporting evidence

4. Identify trials and therapeutic options
- Match genomic profile and diagnosis with treatments and region-specific clinical trials
- Review curated evidence to make final decision on reportability

5. Add oncologist-ready summaries
- Use over 490,000 oncologist-reviewed
variant interpretation comments - Submit rare or novel variants to QIAGEN’s professional
variant interpretation service (optional) - Receive results same day (cases vary)

6. Generate and sign-off on final report
- Build easy-to-understand, customizable reports for oncologists
- Include key details to guide patient treatment, such as variant therapeutic, prognostic and diagnostic relevance and co-occurring interactions
Request a free demo of QCI Interpret for Oncology
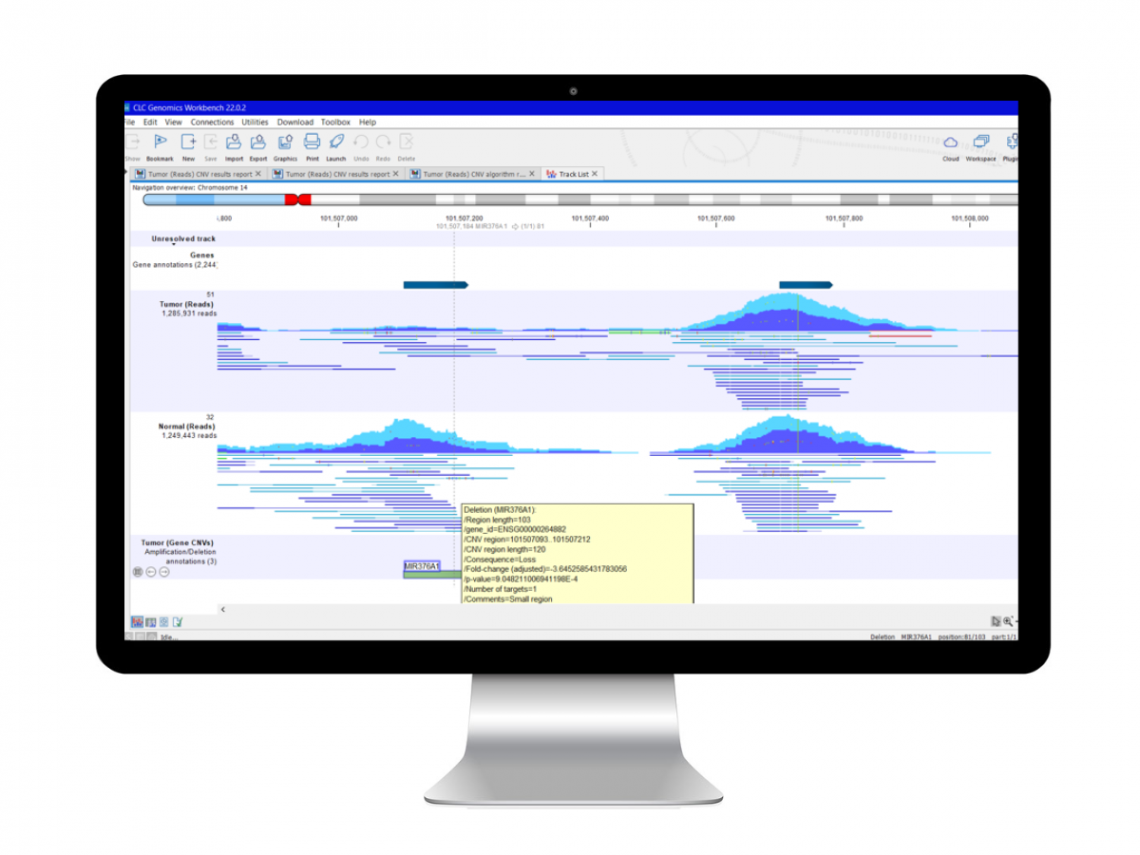
QCI Secondary Analysis
QCI Secondary Analysis is an optional cloud-based service that processes FASTQ files to produce VCF files containing single nucleotide variants (SNV), insertion–deletion mutation (InDel) and structural variant (SV) calls. The module performs quality and adapter trimming, read mapping, deduplication, local realignment, quality control and variant calling and seamlessly connects to QCI Interpret for an integrated and automated FASTQ to report workflow.
- Unrivalled speed -Requires less than 1 minute to process one WESvsample at 35x coverage; can process large comprehensive cancer panels in seconds.
- Lowest cost per sample –Costs less than $0.40 per WGS sample.
- Accurate –Achieves 99% accuracy for more than 90% of the genome.
QCI Interpret for Oncology
QCI Interpret for Oncology is clinical decision support software that combines the unmatched accuracy and consistency of QIAGEN’s proprietary expert (MD/PhD) curation with the superior efficiency of machine curation (AI-powered curation) to enable high-confidence variant interpretation and reporting. The software dynamically computes pathogenicity and actionability based on the AMP/ASCO/CAP or ACMG/AMP guidelines for every variant in over 31,000 cancer types with full transparency. To simplify and accelerate interpretation, users have access to over 320,000 preformulated, oncologist-reviewed variant impact summaries and the in-software option to submit rare or novel variants to QIAGEN’s professional variant interpretation service. Panel- and sequencer-agnostic, QCI Interpret for Oncology can be fully customized to accommodate targered panels, comprehensive genomic profiling, exomes, and genomes.
Software features
Explore the features of QCI Interpret
Use QCI Interpret for Oncology as a variant analysis, interpretation, and decision support software to evaluate somatic genetic variants in the context of professional association guidelines, published clinical cases, clinical trials, and publicly available databases. Quickly retrieve curated variant lists obtained from comprehensive tumor genomic profiling.
Use QCI Interpret for Oncology to group, filter, and prioritize genetic variants from the variant lists. Find actionable mutations in driver genes and match driver alterations with specific drugs allowing personalized therapeutic management. Sort your variants by interpretation type, alteration type, and clinical actionability in search for those that could be used as prognostic and therapeutic biomarkers.
Clinical cases are deeply curated to gather specific evidence for automated computation of an AMP-recommended classification into 4 categories: Tier 1- variants of strong clinical significance (Level of evidence A and B), Tier 2- Variants of potential clinical significance (Level of evidence C and D), Tier 3 –Variants of unknown clinical significance, and Tier 4- Benign or Likely benign variants. For each computed classification the criteria engaged are displayed along with the supporting evidence.
QCI Interpret for Oncology goes beyond genomic descriptive information to include data on clinical impact (diagnostic, prognostic, predictive), matched drugs available, and therapeutic effect. When searching for appropriate therapeutic options, the actual diagnosis is usually used to match treatments and clinical trials. QCI Interpret for Oncology offers the opportunity to search for treatment and clinical trials even in the case of an unknown diagnosis.
The QIAGEN Knowledge Base contains published articles that refer to the specific variants, along with the categorization of the article types: clinical cases, functional studies, drug labels and guidelines, treatment studies, prognostic studies, reviews, and external database reports.
In QCI Interpret for Oncology you can inspect and evaluate the curated data to make a final decision on the pathogenicity/actionability assessment and reportability status (allow the variant to be displayed on the final clinical report). When vetting the criteria in the Assessment window, you can easily add your own criteria for the final variant assessment.
QCI Interpret for Oncology provides expert test interpretation with the updated new world data from basic research and clinical trials. QIAGEN’s goal is to enable customers to generate real-world insights from increasingly large genomic data sets.
QCI Interpret for Oncology enables you to simultaneously search for both single nucleotide variants (SNVs) and copy number variants (CNVs) in each sample. The software provides an integrative view of the small variations together with large exonic indels. To narrow down the list of variants, you can filter and prioritize them according to actionability.
QCI Interpret for Oncology lists the co-occurring variants in each sample. If the mutations occur in the same gene, the software’s “protein view” shows the presence of the mutations, their positions, and their effect on the protein.
QCI Interpret for Oncology identifies and lists co-occurring variants in each clinical sample, providing evidence on the clinical effect with reference to relevant guidelines. The software allows you to filter variants according to genes in which actionable mutations are detected and to visualize the co-mutations that exist in the sample. Users also receive an expert explanation on the clinical effect of the co-occurring mutations with reference to clinical guidelines.
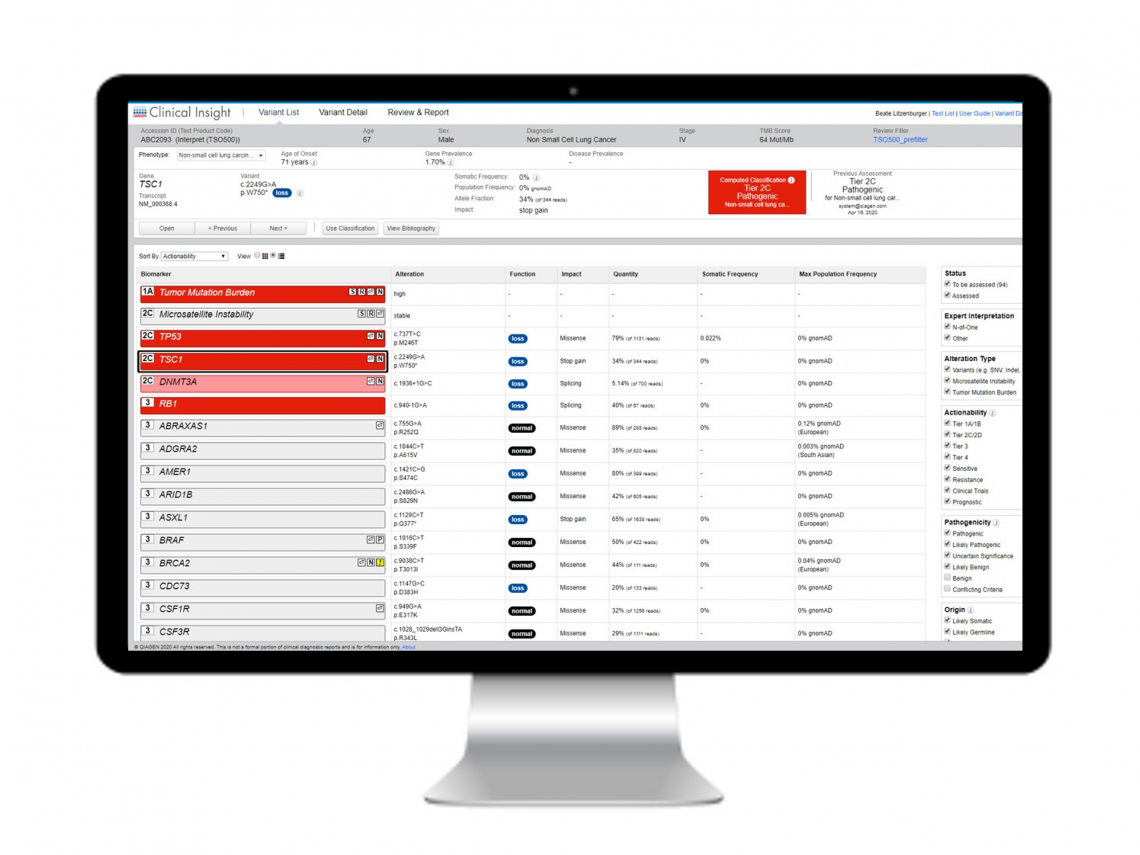
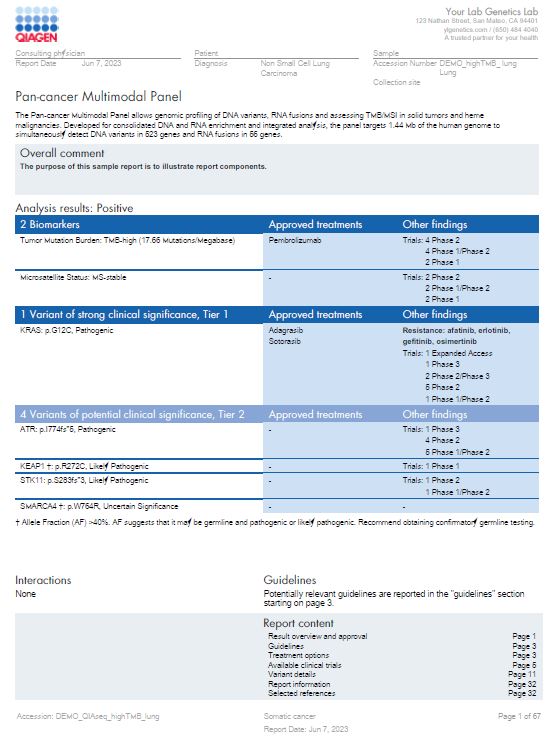
Sample report
QCI Interpret for Oncology sample report
QCI Interpret provides a comprehensive and flexible reporting system that automatically incorporates significant variants, key findings, annotation sources, and interpretation summaries. Reports can be fully customized to meet your lab’s brand and formatting requirements. This sample report is for a pan-cancer multimodal panel and shows results with TMB and MSI biomarkers and a KRAS alteration detected.



Over 3.5 million NGS patient cases interpreted worldwide
Discover why QCI Interpret is one of the most trusted clinical bioinformatics platforms in the world.
Related NGS testing solutions
QCI Precision Insights
Human Somatic Mutation Database
QCI Interpret Oncology Software
QCI Interpret for Oncology
Because oncologists and
their patients rely on you
For molecular pathologists, there are no second chances.
QCI Interpret for Oncology ensures you make the right decisions—faster.
Analyze with Precision. Interpret with Confidence.

The right information in the shortest amount of time
In the NGS era, molecular pathologists have become the clinician responsible for interpreting molecular data and optimizing patient outcomes through the delivery of accurate diagnoses and treatment guidance. Now more than ever, oncologists rely on molecular pathologists to inform and expedite the therapeutic decision-making process. Trusted by clinicians around the world, QCI Interpret for Oncology ensures you have the right information in the shortest amount of time.


>2.5 million patient cases interpreted by QCI Interpret
>6.4 million variants characterized in 18,000 genes
The power of augmented
molecular intelligence
QCI Interpret is a clinical decision support software powered by augmented molecular intelligence that helps clinical labs not only make faster decisions—but the right decisions.
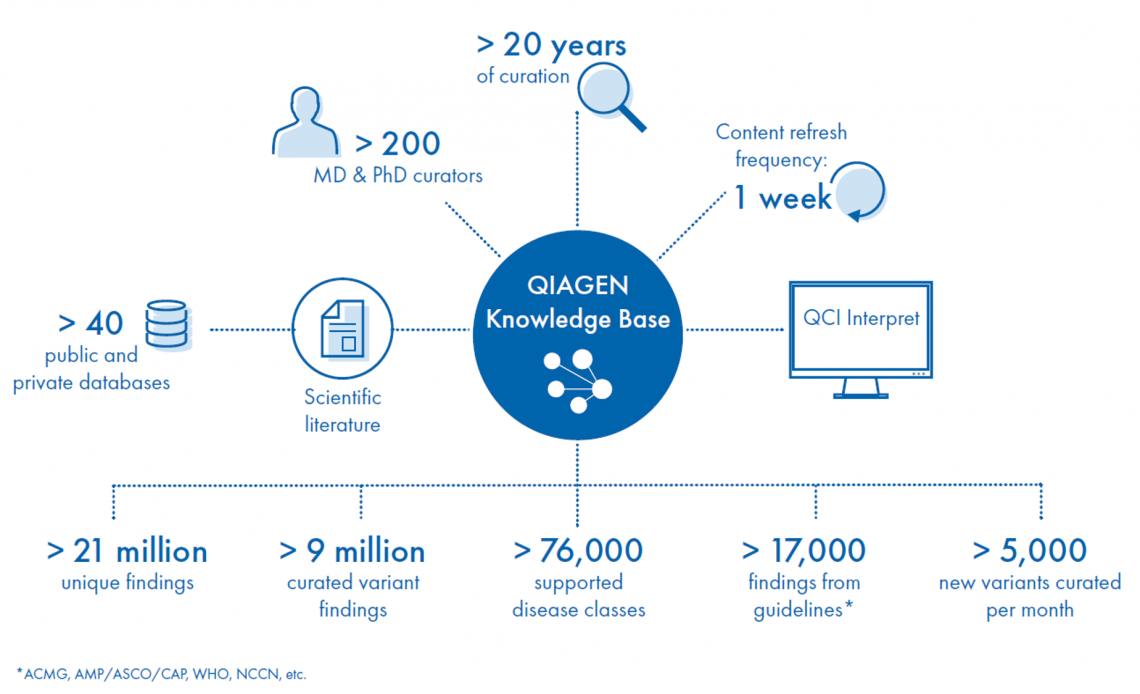
Connected to the exclusive QIAGEN Knowledge Base, the industry’s most comprehensive, manually curated resource that is updated weekly, QCI Interpret for Oncology dynamically computes pathogenicity and actionability based on the AMP/ASCO/CAP or ACMG/AMP guidelines for every variant in over 31,000 cancer types with full transparency. To simplify and accelerate interpretation, users have access to over 320,000 preformulated, oncologist-reviewed variant impact summaries to build custom, patient-specific reports with the latest diagnostic and prognostic information, as well as biomarker-directed therapies and clinical trials.
The content core of QCI Interpret for Oncology, the QIAGEN Knowledge Base is the world’s largest source of globally trusted molecular knowledge. Built manually over 20 years by hundreds of MD- and PhD-level expert curators and augmented by artificial intelligence to rapidly identify, extract, and enhance evidence, the QIAGEN Knowledge Base is unrivalled in breadth, depth, and accuracy. To date, the QIAGEN Knowledge Base has been trusted to analyze and interpret NGS data from over 2.5 million clinical cases worldwide

The QCI Interpret difference
Workflow Agnostic
Special feature: For labs using the Illumina® TruSight Oncology 500 assay, the QCI software solution provides a preconfigured workflow that accepts FASTQ and VCF files.
Fastest Growing Knowledge Base
Compute Any Variant for Any Disease
For every variant in over 31,000 cancer types, QCI Interpret automatically provides a computed AMP/ASCO/CAP classification, computed molecular function, the alteration’s incidence in disease, an oncologist-reviewed interpretation summary, information on hereditary clinical cases, treatment information, including alteration-specific drug sensitivity and resistance, and a list of open and recruiting clinical trials specific to the patient’s geographic region.
Oncologist-Reviewed Variant Summaries
To date, the software provides access to over 320,000 decision-ready interpretive comments to help you accelerate test turnaround time and increase caseload volume.
Interpretation of Co-occuring Mutations
If the mutation occurs in the same gene, the software provides a "protein view" that shows the presence of mutations, their position, and effect on the protein. Users also receive an expert explanation on the clinical effect of the co-occurring mutations with supporting references.
On-demand Interpretation Services
An ideal solution for labs working with rare or novel variants, the on-demand service does the research, curation, and interpretation for you. Any somatic NGS panel can be submitted.
It’s one of our biggest precision oncology events of the year!
May 19 (Part 1) | June 16 & 21 (Part 2)
You’re invited to QIAGEN Digital Insights’ annual Break the Data Bottleneck Summit, a free-to-attend, two-part virtual event exploring how diagnostic labs can lower the barrier to adopt large cancer genomic profiling.
See what's new in the
latest QCI Interpret release
The world's fastest growing knowledge base
QCI Interpret for Oncology contains over 6.4 million precise variants characterized in 18,000 genes.
With over 5,000 new variants added each month, QCI Interpret for Oncology adds a total of 60,000 new variants per year.
Trusted by labs around the world, QCI Interpret for Oncology processes over 15,000 cases each month.
Get exclusive access to evidence curated by variant scientists
Built over 2 decades, the QIAGEN Knowledge Base leverages machine learning to rapidly index journal articles for mutations and human judgment and expertise (Augmented Molecular Intelligence) to ensure accuracy, relevance and context—every catalogued “finding” has been “touched” by a trained scientist.
The QIAGEN Knowledge Base is built on our own comprehensive ontology that uniformly models relationships between different entities, such as the relationship between a variant, the gene that it resides in, and the observed phenotype. The superior structured content ensures consistency and computability.
FASTQ to report within minutes—not hours
QCI Interpret is panel- and sequencer-agnostic and can be used with any secondary analysis platform.
Enabling seamless workflows from FASTQ to report, the software solution supports the interpretation and reporting of targeted panels, comprehensive genomic profiling, clinical exomes and genomes.
For labs using an Illumina® sequencer and Illumina BaseSpace®, raw FASTQ sequencing files can be directly uploaded to QCI Secondary Analysis. For labs using other sequencers, a bulk file uploader tool can be used to upload data to QCI Secondary Analysis.
QCI Secondary Analysis then seamlessly connects to QCI Interpret for Oncology.
The QCI Interpret for Oncology workflow starts with any of the following NGS format files: vcf, zip, gz, tgz, csv, tsv or bz2 file, and it is compatible with any NGS platform. You can choose to upload multiple-sample files for different patients, or multiple single sample files for samples from the same patient. Manually enter metadata to contextualize the uploaded file in the somatic workflow.
Simultaneously upload SNVs and structural variants
In cases when you have separate files for small nucleotide variations (SNVs), copy number variations (CNVs) or fusions and rearrangements for the same patient, upload them at the same time for an integrative view of the small variations together with large exonic indels (see how here).
QCI Interpret for Oncology enables the filtering and prioritization of variants by interpretation type, alteration type, and clinical actionability.
The software uses rules-based approach to automatically compute actionability (Tier 1 to Tier 4) and pathogenicity (Pathogenic to Benign) classifications according to the professional guidelines from the American College of Medical Genetics and Association for Molecular Pathology (ACMG/AMP). Simplify your data for easier analysis by filtering them into several priority levels for gradual review from high priority to low (see how here). Look further into the ones that have been classified by QCI Interpret as clinically actionable (see how here).
Review computed actionability
Actionability and pathogenicity classifications in QCI Interpret are accompanied by clear visibility into the evidence and criteria supporting the classifications. In addition, you can manually add a rationale for each of the rules triggered if desired and adjust the strength of the final assessment if additional data is available (see how here).
Review the lists of treatments and clinical trials
QCI Interpret allows you to search for appropriate therapeutic options by matching the genomic profile and disease diagnosis with the treatments and clinical trials. Combination genotypes are curated from drug labels, guidelines, clinical trials records, and clinical studies. QCI Interpret offers you the opportunity to explore prognostic data, diagnostic data, treatments, and age-matched clinical trials even in the case diagnosis is unknown, or in the case when a match to a specific diagnosis cannot be made.
The treatment/trial is displayed and the Evidence field illustrates the nature of the multiple alteration matches. Look for the clinical trial close to your geographic region and search for drugs that are available in your country (see how here).
Review the literature
QCI Interpret provides expertly-curated extensive bibliography coverage (clinical cases, functional studies, population studies, drug labels and guidelines, treatment studies, prognostic studies, reviews, and external database reports) with multiple lines of evidence linking variants to a disease. The bibliography is categorized by article type for every variant detected and includes all findings that have been curated from the published literature as well as findings that have been imported from specific databases (see how here).
Assess the underlying evidence
Inspect and evaluate the curated data to make a final decision on the actionability assessment and reportability status (allow the variant to be displayed in the final clinical report). Chose the lists of drugs and clinical trials to be listed in the final report. You can easily add your own criteria for the final variant assessment in the Assessment window (see how here).
Leave the heavy-lifting to QIAGEN. On top of accessing over 320,000 decision-ready interpretive comments, you can submit your variants to QIAGEN to receive customized, oncologist-reviewed interpretations and sum-mary comments for every clinically relevant variant detected. An ideal solution for labs working with rare or novel variants, QCI Interpret’s on-demand clinical curation and interpretation services does the research, curation, and interpretation for you, replacing labor intensive processes with automated simplicity.
Generate a final clinical report that is patient-specific and includes clinically relevant variants, interpretations, and references specified throughout the assessment process.
QCI Interpret for Oncology enables rapid report building by providing oncologist-reviewed variant interpretation summaries.
Each clinical report contains the following information:
- Molecular function
- Therapeutic, prognostic, and diagnostic relevance
- Variant interactions, such as effect of co-occurring variants on therapies, drug resistance and sensitivities
• Clinical practice guideline recommendations
• Relevant local recruiting clinical trials
• FDA-approved drug therapies
• Primary literature references
View full sample report here.
Features
Explore QCI Interpret's capabilities
A world of evidence at your fingertips
With QCI Interpret for Oncology, you can be confident that every clinical recommendation you make is backed by the latest peer-reviewed publications, clinical practice guidelines, FDA therapeutics, and open clinical trials, all vetted by M.D. and Ph.D.-level expert curators who do the reading for you.
Use QCI Interpret for Oncology as a variant analysis, interpretation, and decision support software to evaluate somatic genetic variants in the context of professional association guidelines, published clinical cases, clinical trials, and publicly available databases. Quickly retrieve curated variant lists obtained from comprehensive tumor genomic profiling.
Use QCI Interpret for Oncology to group, filter, and prioritize genetic variants from the variant lists. Find actionable mutations in driver genes and match driver alterations with specific drugs allowing personalized therapeutic management. Sort your variants by interpretation type, alteration type, and clinical actionability in search for those that could be used as prognostic and therapeutic biomarkers.
Clinical cases are deeply curated to gather specific evidence for automated computation of an AMP-recommended classification into 4 categories: Tier 1- variants of strong clinical significance (Level of evidence A and B), Tier 2- Variants of potential clinical significance (Level of evidence C and D), Tier 3 –Variants of unknown clinical significance, and Tier 4- Benign or Likely benign variants. For each computed classification the criteria engaged are displayed along with the supporting evidence.
QCI Interpret for Oncology goes beyond genomic descriptive information to include data on clinical impact (diagnostic, prognostic, predictive), matched drugs available, and therapeutic effect. When searching for appropriate therapeutic options, the actual diagnosis is usually used to match treatments and clinical trials. QCI Interpret for Oncology offers the opportunity to search for treatment and clinical trials even in the case of an unknown diagnosis.
The QIAGEN Knowledge Base contains published articles that refer to the specific variants, along with the categorization of the article types: clinical cases, functional studies, drug labels and guidelines, treatment studies, prognostic studies, reviews, and external database reports.
In QCI Interpret for Oncology you can inspect and evaluate the curated data to make a final decision on the pathogenicity/actionability assessment and reportability status (allow the variant to be displayed on the final clinical report). When vetting the criteria in the Assessment window, you can easily add your own criteria for the final variant assessment.
QCI Interpret for Oncology provides expert test interpretation with the updated new world data from basic research and clinical trials. QIAGEN’s goal is to enable customers to generate real-world insights from increasingly large genomic data sets.
QCI Interpret for Oncology enables you to simultaneously search for both single nucleotide variants (SNVs) and copy number variants (CNVs) in each sample. The software provides an integrative view of the small variations together with large exonic indels. To narrow down the list of variants, you can filter and prioritize them according to actionability.
QCI Interpret for Oncology lists the co-occurring variants in each sample. If the mutations occur in the same gene, the software’s “protein view” shows the presence of the mutations, their positions, and their effect on the protein.
QCI Interpret for Oncology identifies and lists co-occurring variants in each clinical sample, providing evidence on the clinical effect with reference to relevant guidelines. The software allows you to filter variants according to genes in which actionable mutations are detected and to visualize the co-mutations that exist in the sample. Users also receive an expert explanation on the clinical effect of the co-occurring mutations with reference to clinical guidelines.
From our customers
Success stories
Quest Diagnostics
Protean BioDiagnostics
Unilabs Switzerland



Contact us
See the content we
have for your variants
Want to see what content QCI Interpret for Oncology can provide for your variants? Please complete the form below and our experts will show you the depth of content that our software provides for your specific variants.