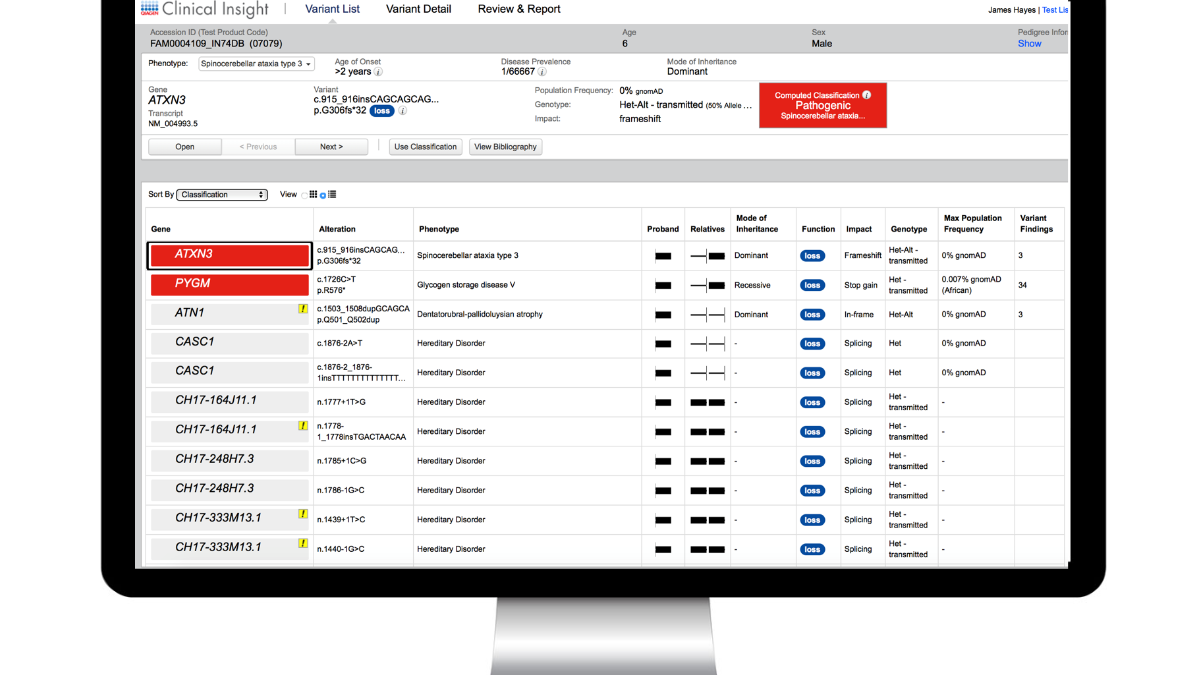
In its latest release, QCI Interpret advances market-leading NGS interpretation software with artificial intelligence-driven insights to enable clinical exome completeness
We are pleased to announce that the Summer 2023 Release of QCI Interpret, QIAGEN’s decision support software platform for the annotation, classification, and reporting of somatic and germline variants, is now available.
Expanding on the software’s current capabilities, the QCI Interpret Summer 2023 Release extends its market-leading, unrivalled content with further advancements in Artificial Intelligence (AI) to enable clinical exome completeness, enhanced phenotype driven ranking, and improved literature searches.
QCI Interpret Summer 2023 Release highlights
- QCI Interpret leverages artificial intelligence (AI) to expand the bibliography: To provide broader coverage of the clinical exome, AI-derived literature references have been integrated into the Bibliography section. The new Source icons, “Context” disease details, and “Show Phenotype-Related References Only” filter provide full control over the use of these automatically extracted references.
- Enhanced, AI-trained phenotype-driven ranking: Variant-specific qualities are now taken into account beyond the similarity of the gene–disease phenotypic spectrum to patient symptoms by providing an improved PDR score ranking of candidate variants for rapid clinical exome review.
- Improved ACMG PVS1 criteria with the modified strength workflow and informative notices: ClinGen’s PVS1–modified strength recommendations are now applied to improve the ACMG classification of loss of function variants. PVS1 notices indicate the specific decision tree branch applied.
- Gene transcripts with MANE select status are now the default: QCII embraces this community effort to standardize clinical transcripts for reporting. “MANE Select” and “MANE Plus Clinical” labels appear in the variant viewer dropdown, in the “Effect on Protein” section of the Variant Details page.
- More precise structural variant filtering: The Confidence filter now supports thresholds for copy number gains, losses, fusions, and rearrangements. A “gene–associated structural variants” filter enables retention of structural variants that are partly in non-coding gene regions.
- Enhanced accuracy of the Somatic reporting policy: The 1–year expiration was removed for user set assessments in Somatic reporting policies v1 and v2. User assessments are now automatically re-reported indefinitely in new cases for all reportability types, including artifacts.
- New workflows for streamlined report revisioning and labeling: A new Label Report button on the Review and Report page enables the addition (or removal) of a report label including Addendum, Amendment, Correction, or custom text. The new Addend Report button on signed out tests creates a copy, that is labeled with Addendum, to facilitate issuing an updated report. API enhancements enable a new report to replace an existing one (correction) or creation of an amended/addended report copied from an existing report with updated information (such as patient or physician information) provided via the API. The report label can be displayed on customized reports. A fee may apply to update existing customized report templates.
- Refined handling for multiple fusions with the same first breakpoint: On the Review & Report page, multiple fusions with the same first breakpoint location and gene pair were previously collapsed and shown as a single variant. Now the specific fusion breakpoint pair(s) are maintained on the Review & Report page for additional clarity.
- Advanced handling of EGFRvIII: EGFR exon 2–7 deletion and EGFR e1:e8 rearrangements are now handled the same for identifying treatments and clinical trials. Non-specific EGFR targeted treatments and trials are no longer automatically reported for EGFRvIII.
- Increased privacy and protection: For security and data privacy, idle sessions now result in automatic logout after 30 minutes of inactivity. A notice appears minutes before a session will be ended which permits the session to be extended.
Understanding QCI Interpret's manual and AI curation process
The latest release of QCI Interpret for Hereditary includes AI enhanced coverage of thousands of rare disease genes. The below graphic illustrates the improved curation process for QCI Interpret, showing how all content is initially extracted using AI and machine learning, the top 1,000 routinely tested genes undergo human-certified curation, and all content undergoes rigorous quality control to ensure consistency and accuracy (Figure 1).
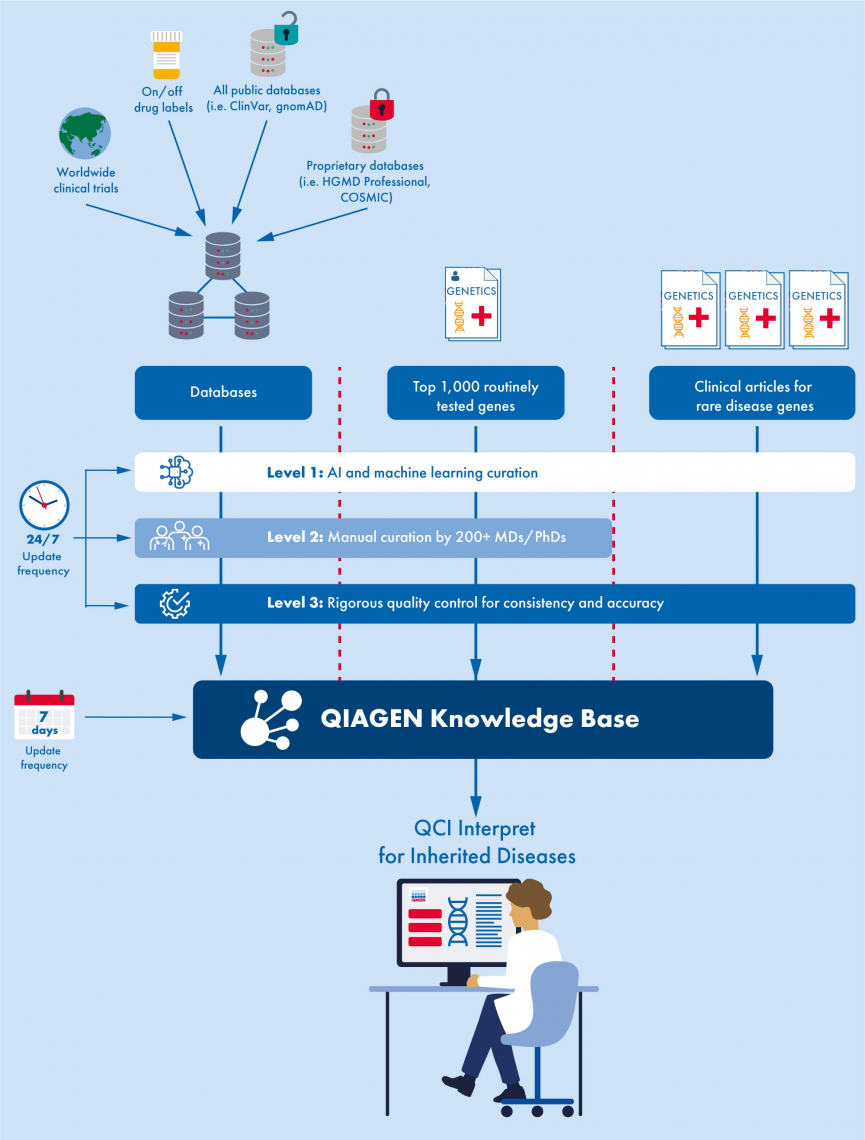
Figure 1. The comprehensive curation process for QCI Interpret for Hereditary.
About QCI Interpret
QCI Interpret is a clinical decision support software platform for the annotation, classification, and reporting of actionable alterations from NGS data for oncology and hereditary disease applications. Using AI-enabled literature searches and expertly curated content from the QIAGEN Knowledge Base, QCI Interpret applies a rules-based approach to automatically compute pathogenicity classifications (Pathogenic to Benign) and actionability classifications (Tier 1 to 4) for each alteration according to professional guidelines from ACMG/AMP and AMP/ASCO/CAP, respectively.
Pathogenicity and actionability classifications in QCI Interpret are accompanied by clear visibility into the criteria and evidence supporting the classifications. This workflow starts with a variant call format (VCF) file, so it is compatible with the output from any NGS platform. The final report includes the alterations, interpretations, and references specified throughout the assessment process, which has customizable automation capabilities allowing for streamlined clinical decision support workflows.