


















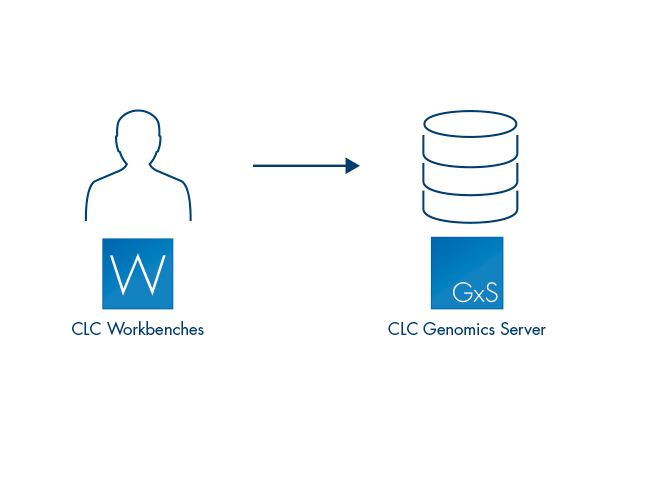
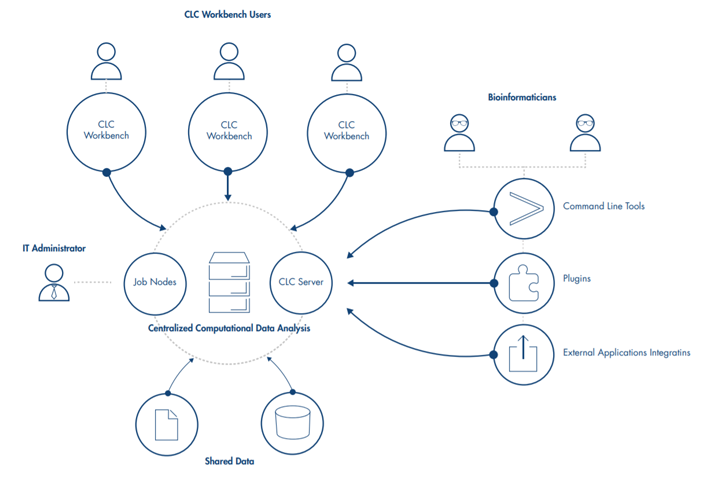
QIAGEN CLC Genomics Server is a multi-user, enterprise solution for high-throughput sequencing analysis, designed for use on a central compute cluster or server. Along with a strong bioinformatics analysis offering, it can handle data volumes beyond the capacity of desktop systems and effectively manages the submission of many jobs via its own queuing system or through submission of jobs to a third party grid scheduler. It is designed to be easy and intuitive to use: a graphical interface for administration is included, and jobs can be submitted to the QIAGEN CLC Genomics Server through the user friendly, graphical menu system of the QIAGEN CLC Genomics Workbench or by using the command line.

After the release of guidelines like ICH Q5A(R2) and 21 CFR Part 11, more biopharma companies are switching to next-generation sequencing (NGS) for viral safety.
QIAGEN CLC Genomics Server offers an auditable, customizable environment that streamlines your compliance and let your company navigate these regulatory changes with ease.
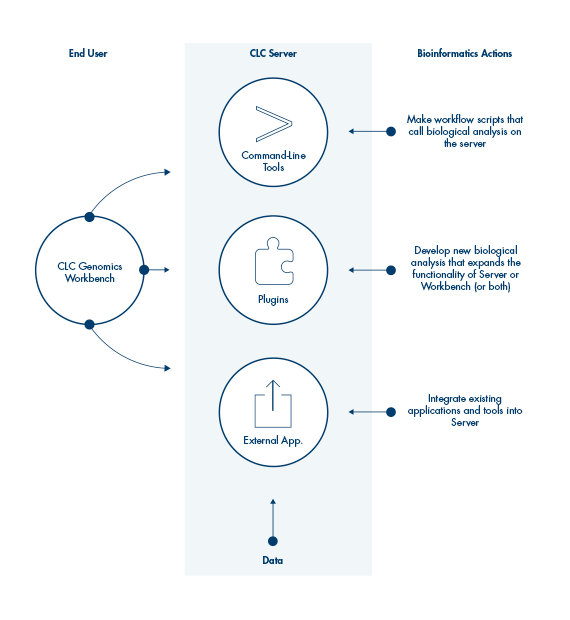
QIAGEN CLC Genomics Server supports import and export of major bioinformatics file formats, such as fastq, fasta, BAM, VCF, BED and others, and provides bioinformatics tools for the analysis of next generation sequencing data in many application areas. Plugins further expand its functionality, including supplying ready-to-use workflows for application areas such as biomedical analysis, microbial genomic analysis, and QIAseq panel data analysis.
Workflows on QIAGEN CLC Workbenches can be executed on the QIAGEN CLC Genomics Server, or they can be exported and installed directly on the QIAGEN CLC Genomics Server, allowing the same workflow, with the same settings, to be employed by multiple users.

QIAGEN CLC Genomics Server can be run on computer clusters either as a QIAGEN job node setup or by integrating with an existing grid setup via a DRMAA interface. We have verified our grid integration on the following systems:

We frequently release updates and improvements such as new functionalities, bug fixes or plugins. A full list of recent changes are listed on the latest improvements page.
Analyses on the QIAGEN CLC Genomics Server can be initiated using a QIAGEN CLC Genomics Workbench or the CLC Server Command Line Tools.