Rapidly and efficiently annotate genetic variants from high-throughput sequencing data and predict their functionalities.
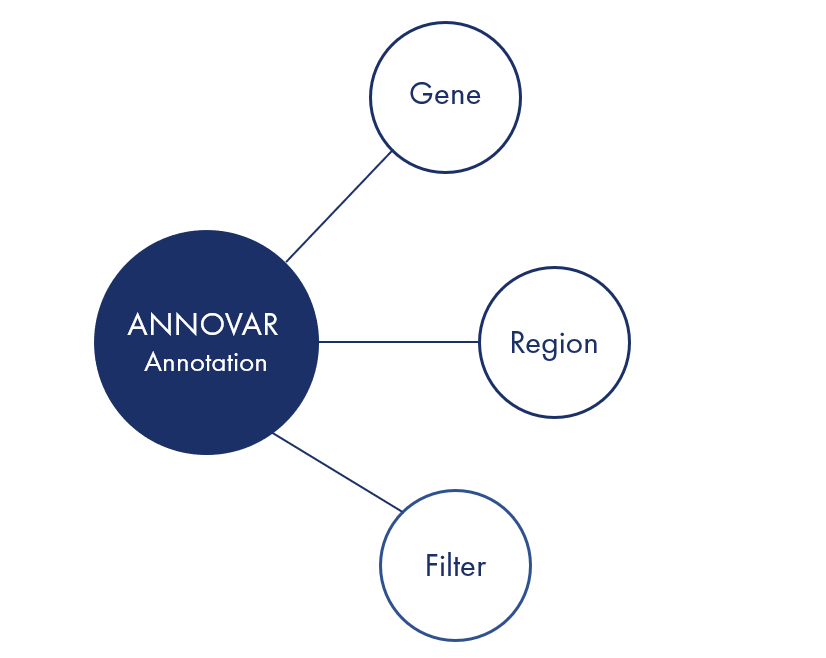
ANNOVAR is a fast and flexible Perl command line program that enables high-throughput functional annotation and filtering of genetic variants from next-generation sequencing (NGS) data. ANNOVAR allows researchers to annotate a whole genome in under 4 minutes and can handle hundreds of genomes per day on a standard desktop PC.




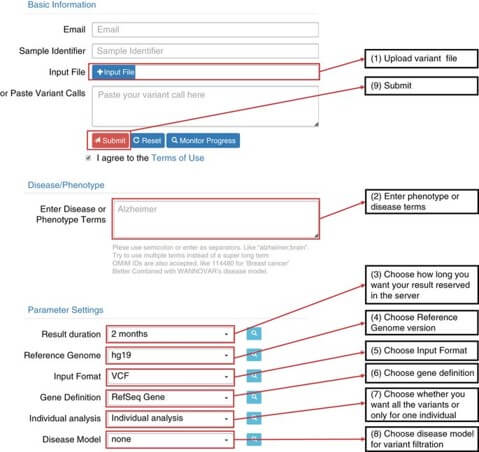
To facilitate convenient and fast access to ANNOVAR for researchers who prefer a graphical user interface, wANNOVAR provides web-based access to the most commonly used functionalities of ANNOVAR for human genomes.
By visiting the wANNOVAR website, users can directly upload their variant files and obtain their results back via web interface.
In addition, wANNOVAR implements phenotype-based variant prioritization, which is helpful in scenarios in which a sample's specific phenotype or disease information is available and may help identify causal variants.