

















A new functionality in QIAGEN® Biomedical Genomics Workbench can be used in conjunction with Ingenuity® Variant Analysis™ to identify causal variants, which disrupt protein/drug binding or have an impact on the protein 3D structure. The user gains an insight not only if a variant is probably causal, but also why.
The new tool, Link Variant to 3D Protein Structure, together with the new protein 3D viewer, offers a quick method for investigating the likely impact of a putative causal variant on the protein 3D structure. For each variant, the Biomedical Genomics Workbench combs through known protein structure information in the PDB database to predict the effect on structure, even for novel mutations. The tool also allows users to generate hypotheses for the effect of a mutation based on what part of the protein it changes. By eliminating the need to query a publicly available protein structure database for each mutation and then build a homology model, this tool dramatically accelerates a user’s ability to find the likely structural effect of DNA and RNA changes.
To show its effectiveness on a rare disease case, we went back to one of our favorite Ingenuity Variant Analysis data sets, the TGF-β3 data set generated by Dr. Hugh Rienhoff in his analysis of his daughter’s DNA. We loaded the causative mutation into Biomedical Genomics Workbench and used the new functionality to find out why the mutation in TGF- β3 is likely the disease causing one. In less than two minutes, we got the protein structure, which clearly illustrates how the mutation induces an amino acid change that breaks a disulfide bond and clashes with the surrounding protein structure. This suggests that this protein, which is probably non-functional, is damaged in a region required in the assembly of the larger protein complex.
These findings were experimentally validated by Dr. Rienhoff.
Pictures from the 3D viewer in the workbench. The protein chain affected by the mutation is shown in blue ribbon representation.
The leftmost picture shows the protein reference structure. Three disulfide bridges establishing a highly structured part of the protein are shown in sticks, and the cysteine being mutated is labeled (Cys409).
The rightmost picture shows the mutated amino acid (Tyr409) in space-filling, to illustrate how the increased size of the side chain will not fit inside the rigid structure. The amino acid is colored to show clashing atoms in red.
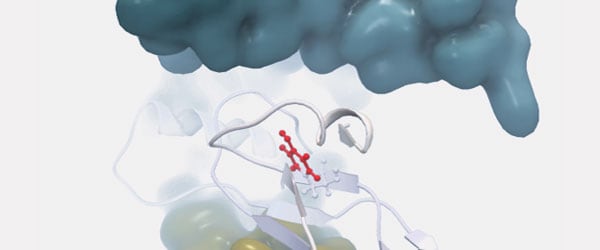
Picture from the 3D viewer in the workbench. The protein chain affected by the mutation is shown in ribbon representation and the amino acid (Tyr409) resulting from the mutation is shown in ball-n-sticks and colored to show clashing atoms in red. Other protein chains from the larger protein complex are shown as molecular surfaces (blue and yellow). The mutation will disrupt an otherwise very stable part of the protein, forming an interface to the protein chain shown in blue surface.
Read more about Dr. Rienhoff’s case study.