QIAGEN Discovery Bioinformatics Services offer custom solutions that help translate discovery from your data. This includes tailored plugin development for QIAGEN CLC Genomics Workbench and QIAGEN CLC Genomics Server, helping to extend and customize the feature sets of these NGS analysis solutions.
The QIAGEN Antibody Analysis Suite is one of the custom solutions developed by the QIAGEN Discovery Bioinformatics Services team. This automated, high-throughput tool enables a detailed analysis of antibody sequences and includes the ability to customize to ensure that it fits the exact needs of specific workflows. The Antibody Analysis Suite is used for the characterization of antibody repertoires involved in the discovery and development of therapeutic and diagnostic antibodies and often includes DNA sequence analysis. In vitro display and selection, for example, requires sequence analysis at several steps in the process. First, to test initial library diversity and quality, and subsequently to ensure proper selection and thorough characterization of final clones. The QIAGEN Antibody Analysis Suite provides a tool for detailed analysis that:
- Accelerates the research process
- Removes manual and error-prone steps
- Is user-friendly and integrated
- Can be customized to match specific needs
The Antibody Analysis Suite is a collection of tools that can be fully integrated with the powerful QIAGEN CLC Genomics bioinformatics platform, seamlessly providing endless options for further up- or downstream analysis. The Antibody Analysis Suite can help you:
- Automate and standardize your workflow for heavy and light chain annotation with QC reports
- Build an internal database
- Provide detailed reports and visuals of gene segment usage and mutations with annotation of antibody-specific features
- Automatically annotate antibody heavy and light variable domains and alignments of both DNA and amino acid sequences to proprietary or public databases of germline DNA sequences, or rearranged variable domain sequences
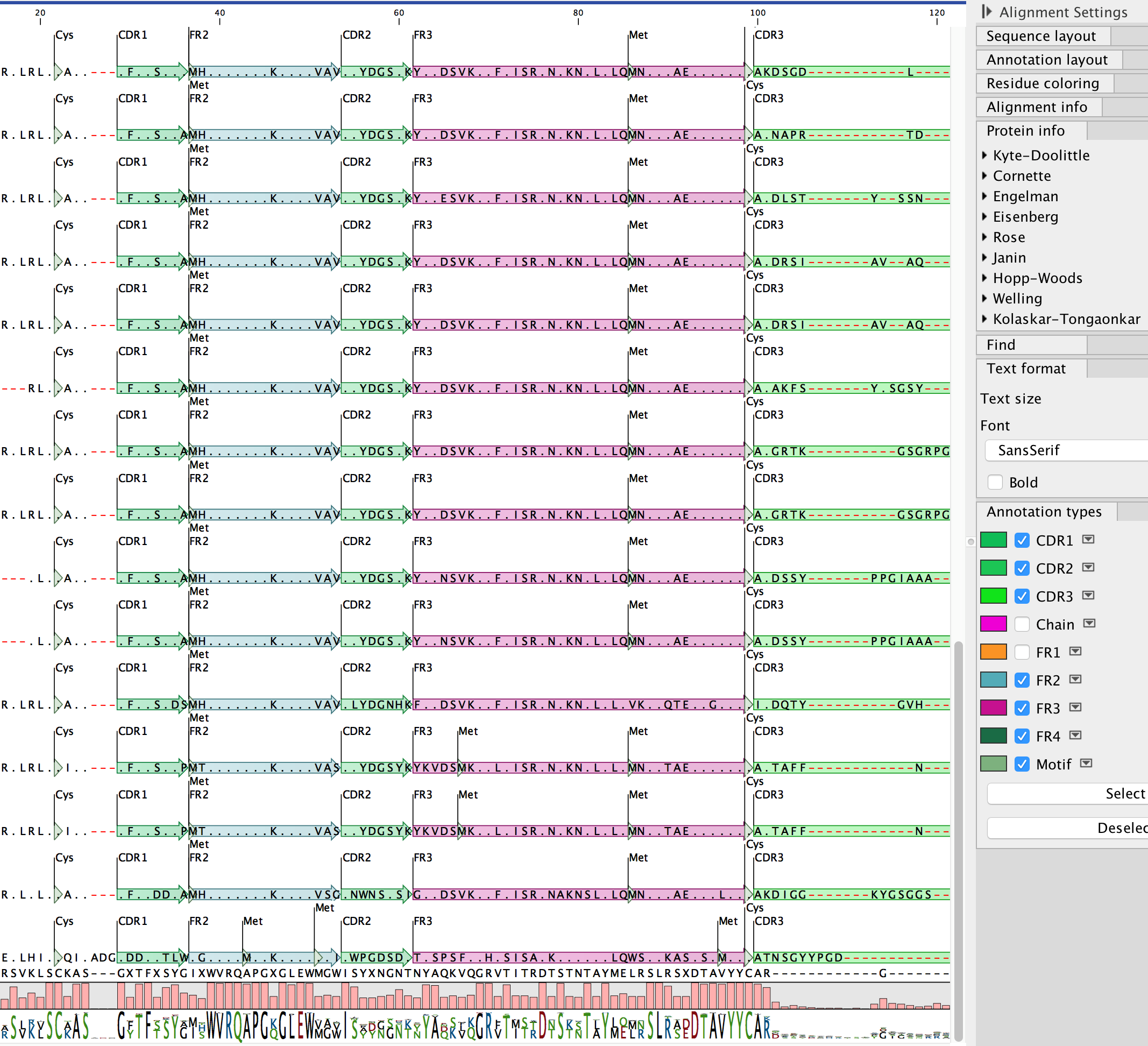
- Understand details regarding V, D and J allele usage and mutations found compared to these; junction sequences; FR and CDR region lengths and sequences, presented in tables and visual displays for further investigation, such as multiple alignments across selected input sequences
- Efficiently identify antibody clones per sample
- Create phylogenetic visualizations and dendrograms
- Perform robust phylogenetic analysis framework that supports metadata-based visualization, as well as sequence clustering
- Analyze results from protein and DNA samples, in parallel
- Determine V, D and J allele usage: Each antibody VH or VL domain is compared to a database of germline sequences of V, D and J alleles, to identify the best hit for each of those gene segments. The alignment of the V segment to its closest germline V segment is displayed in one of many customizable, graphical views
- Perform multiple amino acid alignments: The VH and VL domains of each translated antibody sequence are aligned to a collection of pre-existing, annotated heavy and light domains
- Annotate VH and VL domains: Each sequence is annotated with antibody-specific features, including CDR regions, junction region, specific V, D and J segments and numerous combinations of these
- Quality control your sequences: Each sequence will be checked in terms of frame-shifts and stop codons, as well as missing features, such as conserved amino acids, segments or other motifs
- Group and assemble multiple sequence reads per antibody clone: If an antibody sequence is composed of multiple underlying DNA sequence reads, these can be grouped and assembled prior to analysis
- Create in-house comparisons and databases: Capabilities to make private BLAST databases and protein alignments, such that all analyses can be performed in-house and with proprietary or downloaded public data
- Fully supported to run locally, on servers or cloud environments
Visit us here for more information and request a consultation from our experts today!