

















QCI Interpret Translational's latest software update comes with several new features, including a new workflow for comprehensive cancer panels and greater variant coverage for labs using the GRCh38 reference genome
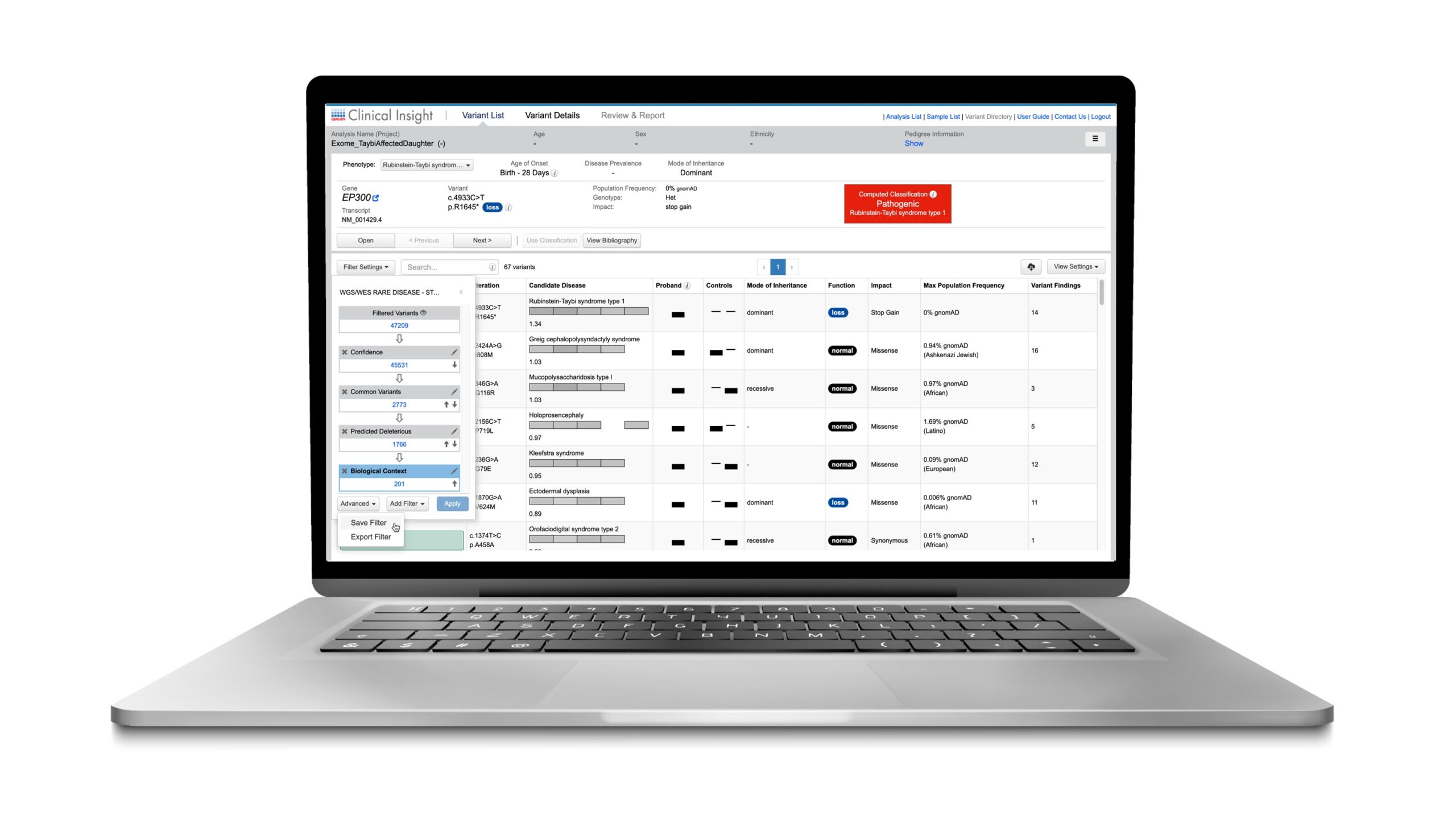
We are pleased to announce that the Summer 2022 Release of QCI Interpret Translational, QIAGEN’s web-based software application for the annotation, classification, and research of variants from next generation sequencing (NGS) data in genomic laboratories, is now available. Expanding on the software’s current capabilities, the QCI Interpret Translational Summer 2022 Release brings new workflows, variant content and functionality improvements.
QCI Interpret Translational is a web-based software application for the annotation, classification, and research of variants from next generation sequencing (NGS) data in genomic laboratories. Using augmented molecular intelligence and expertly curated content from the QIAGEN Knowledge Base, QCI Interpret Translational uses evidence-based approaches to automatically compute pathogenicity classifications (Pathogenic to Benign) and actionability classifications (Tier 1 to 4) for each alteration according to the 2015 professional guidelines from the American College of Medical Genetics and Association for Molecular Pathology (ACMG/AMP) [1] and the 2017 guideline from the Association for Molecular Pathology, American Society of Clinical Oncology, and College of American Pathologists (AMP/ASCO/CAP) [2], respectively.
Pathogenicity and actionability classifications in QCI Interpret Translational are accompanied by clear visibility into the criteria and evidence supporting the classifications. This workflow starts with a variant call format (VCF) file, so it is compatible with the output from any NGS platform. The final analysis is sample-specific and includes candidate causal variants, their deep annotations, interpretations, and references specified throughout the assessment process. The assessment process also has customized automation allowing for even more streamlined variant research workflows.
QCI Interpret Translational helps research labs:
Learn more about QCI Interpret Translational here.