

















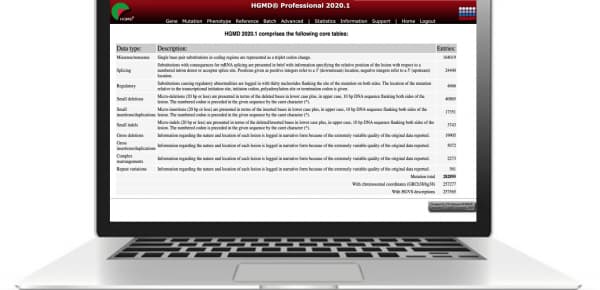
The Spring 2020 Release of the Human Gene Mutation Database (HGMD) Professional is available, expanding the world’s largest collection of human inherited disease mutations to 282,895 entries–that’s 7,179 more than the previous release.
For over 30 years, HGMD Professional has been used worldwide by researchers, clinicians, diagnostic laboratories and genetic counselors as an essential tool for the annotation of next-generation sequencing (NGS) data in routine clinical and translational research. Founded and maintained by the Institute of Medical Genetics at Cardiff University, HGMD Professional provides users with a unique resource containing expert-curated mutations all backed by peer-reviewed publications where there is evidence of clinical impact.
Whether searching for an overview of known mutations associated with a particular disease, interpreting clinical test results, looking for the likely causal mutation in a list of variants, or seeking to integrate mutation content into your custom NGS pipeline or data repository—HGMD is the defacto-standard repository for heritable mutations that can be adapted to a broad range of applications.
detailed mutation reports
new mutation entries in 2019 alone
summary reports listing all known
inherited disease mutations
HGMD is powered by a team of expert curators at Cardiff University. Data are collected weekly by a combination of manual and computerized search procedures. In excess of 250 journals are scanned for articles describing germline mutations causing human genetic disease. The required data are extracted from the original articles and augmented with the necessary supporting data.
The number of disease-associated germline mutations published per year has more than doubled in the past decade (Figure 1). As rare and novel genetic mutations continue to be uncovered, having access to the latest scientific evidence is critical for timely interpretations of next-generation sequencing (NGS) data.

View the complete HGMD Professional statistics here.
Read more about the importance of having access to the most up-to-date and comprehensive database for human disease mutations in our white paper.
HGMD Professional helps clinical testing labs analyze and annotate next-generation sequencing (NGS) data with current and trusted information. Unlike other mutation databases, HGMD mutations are all backed by peer-reviewed publications where there is evidence of clinical impact.
To get the most out of your HGMD Professional subscription, visit our Resources webpage for case studies, technical notes, and video tutorials. Or hear from Peter Stenson, manager of HGMD, in an on-demand webinar on how HGMD has empowered a generation of geneticists for precision medicine here.
Or hear from Peter Stenson, manager of HGMD, in an on-demand webinar on how HGMD has empowered a generation of geneticists for precision medicine here.
An updated version of ANNOVAR is also available.
Learn more about how ANNOVAR can be used with HGMD for variant annotation. Watch a recorded webinar featuring ANNOVAR here.
The Genome Trax™ 2020.1 is now available. Updated tracks have been released with HGMD 2020.1 content for all HGMD-related tracks. Additional major updates include TRANSFAC® release 2020.1, and PROTEOME™ release 2020.1.
For labs looking to generate clinician-grade reports for germline or somatic NGS testing, QIAGEN Clinical Insight (QCI) Interpret reproducibly translates highly complex NGS data into standardized reports using current clinical evidence from the QIAGEN Knowledge Base, which consists of over 40 public and proprietary databases, including HGMD Professional.
Click here for a free demonstration of QCI Interpret.