

















Software Version: 4.1.20160615
Ethnic group specific allele frequency information in Allele Frequency Community
In addition to variant composite frequency, users can now filter against East Asian, South Asian, African (American), European, and Hispanic sub-population frequencies in the Common Variants filter.
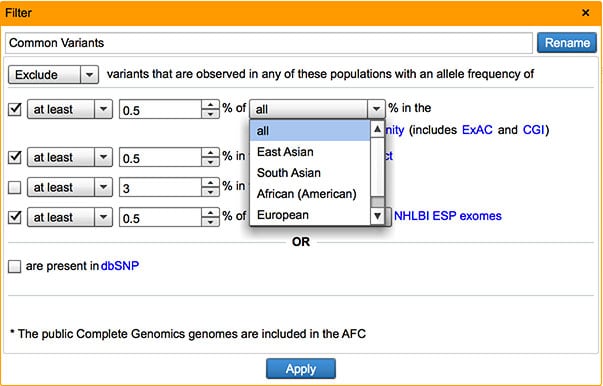
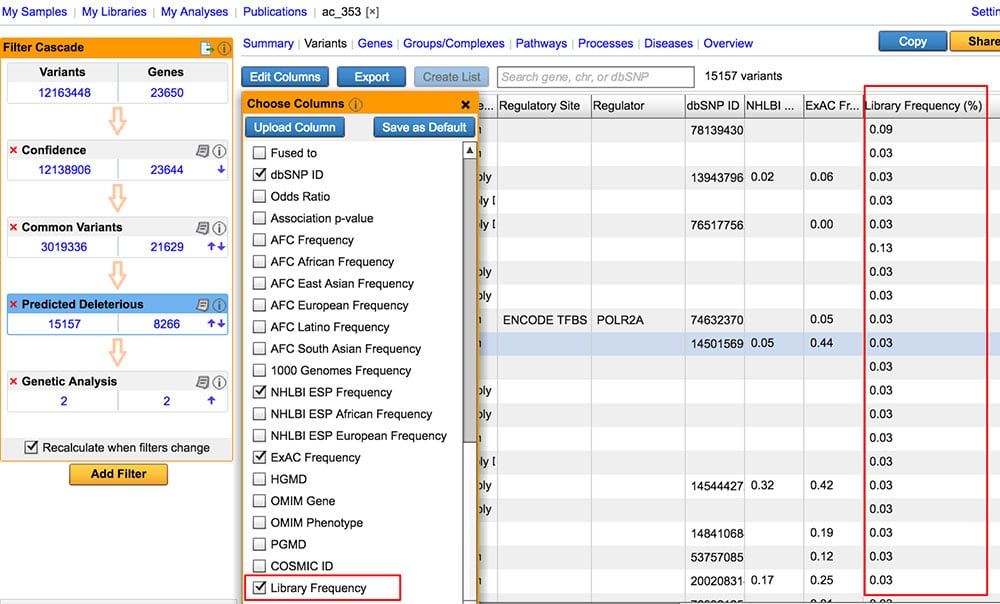
Compute allele frequencies from user-defined collection of samples
Empower users to establish their own “baseline” by computing frequencies based on a collection of samples (“My Libraries”), and annotating variants in a future analysis with library frequencies.
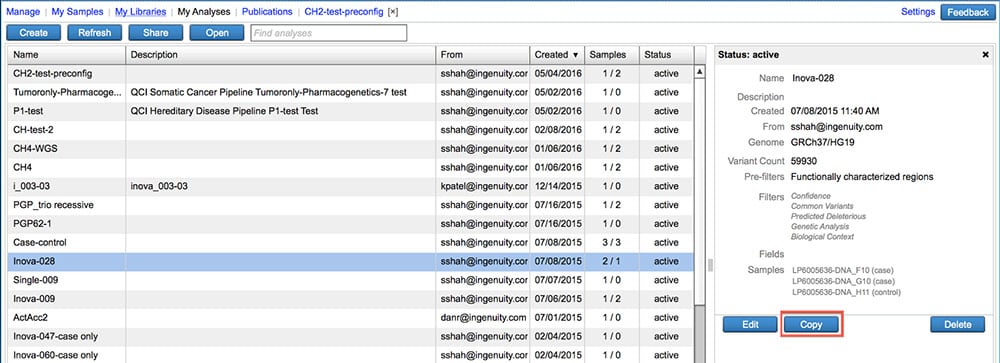
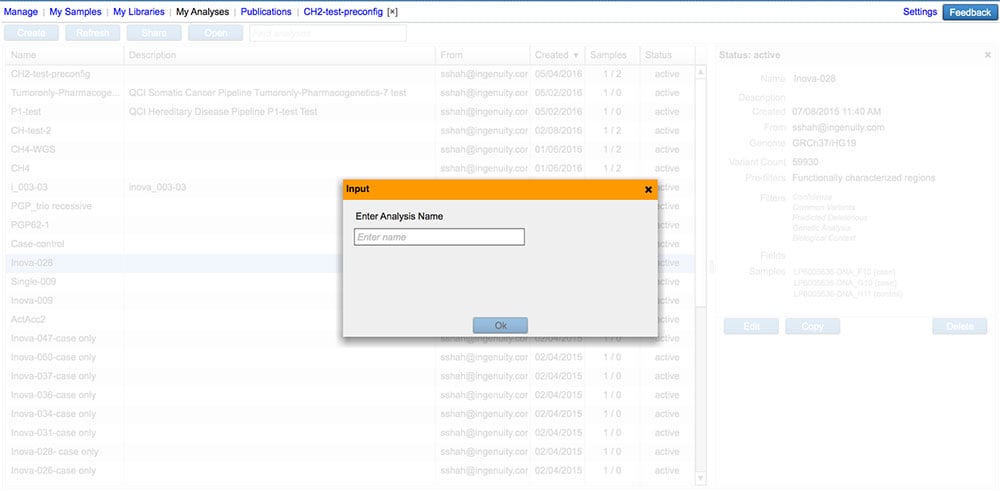
Make a copy of an existing analysis
Carry over analysis filter settings, samples, and sample metadata into a new analysis.
Improvements
Ingenuity Variant Analysis version 4.1.20160615
Content versions: CGI 54 Genomes (Version2.0), SIFT (2016-02-26), EVS (ESP6500SI-V2), Allele Frequency Community (2016-06-16), JASPAR (2013-11), Ingenuity Knowledge Base (Idris 160423.001), Vista Enhancer (2012-07), BSIFT (2016-02-26), TCGA (2013-09-05), PolyPhen-2 (v2.2.2), 1000 Genome Frequency (phase3v5b), Clinvar (2016-03-01), COSMIC (v76), ExAC (0.3), HGMD (2016.1), PhyloP (2019-11), DbSNP (146), TargetScan (6.2)