

















Discovery of new genes implicated in hereditary diseases or cancer progression is challenging and advancing rapidly with an increase in the amount of cohort data to analyze from next generation sequencing (NGS) data to associations between gene variants and diseases, and finally to biological mechanisms of these gene variants. Combining QIAGEN CLC Genomics Workbench, QCII-T, and IPA, scientists can analyze sequencing data obtained from a variety of NGS technologies, annotate gene variants using the world’s most comprehensive and up-to-date curated scientific evidence, and find biological connections in gene variants with manually curated scientific findings. Get valuable and reliable insights for your research project and speed up your discoveries by using a streamlined NGS analysis workflow.
In this webinar, attendees will have the opportunity to:
This year at the 2024 Association for Molecular Pathology (AMP) Annual Meeting, QIAGEN will showcase our barrier-breaking Sample to Insight Oncology solution. Combining targeted DNA and multimodal pan-cancer panels, one of the fastest and cheapest secondary analysis solutions in the market, and industry-leading variant interpretation and reporting software trusted to analyze and interpret over 4 million NGS patient test cases worldwide, our Sample to Insight Oncology solution is revolutionizing precision oncology testing.
Lean more and schedule a VIP meeting with our experts at AMP 2024 here.
This year at the 2024 American Society of Human Genetics (ASHG) Annual Meeting, QIAGEN will showcase our Sample to Insight solutions for human genetics and inherited disease applications. From digital PCR technologies and actionable exome panels to one of the fastest and cheapest secondary analysis solutions in the market and industry-leading variant interpretation and reporting software trusted to analyze and interpret over 4 million NGS patient test cases worldwide, our Sample to Insight solutions are revolutionizing genetic testing.
Learn more and schedule a VIP meeting with our experts at ASHG 2024 here.
This year at the 2024 Precision Medicine Forum Berlin, QIAGEN will showcase our barrier-breaking Sample to Insight Oncology solution. Combining targeted DNA and multimodal pan-cancer panels, one of the fastest and cheapest secondary analysis solutions in the market, and industry-leading variant interpretation and reporting software trusted to analyze and interpret over 4 million NGS patient test cases worldwide, our Sample to Insight Oncology solution is revolutionizing precision oncology testing.
Learn more and register here.
QIAGEN will be at the 36th European Congress of Pathology (ECP) in Florence from September 7-11, 2024, in Florence, Italy.
At the event, be sure to check out our poster that we are presenting in collaboration with a leading pharmaceutical company showing how our clinical decision support software, QCI Interpret, provides rapid and accurate interpretation of gBRCA genes in a clinical lab setting. In the study, 10 partner labs were asked to assess 48 retrospective analyses of BRCA variants and compare their variant classifications with the automated classifications computed by QCI Interpret. Results showed QCI Interpret was concordant with 93% of all cases.
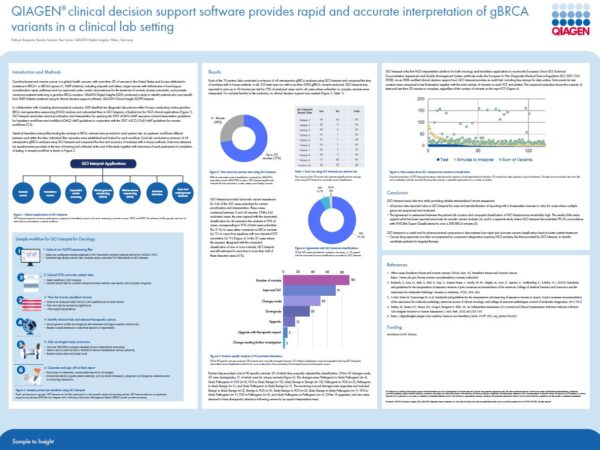
Featured Poster: Spadolini Pavilion (Upper Floor)
Molecular Pathology Session | September, 10, 2024, 9:30–10:30 a.m.
Poster number: 009. Title: “QIAGEN clinical decision support software provides rapid and accurate interpretation of gBRCA variants in a clinical lab setting”
Join us for three 60-minute training sessions for QIAGEN Ingenuity Pathway Analysis (IPA).
In this training, you’ll learn how to:
Tuesday July 9 – Session#1: 1pm ET/10am PT
Wednesday July 10 – Session#2: 1pm ET/10am PT
Thursday July 11 – Session#3: 1pm ET/10am PT
If you are able to attend, then we will have additional scientists on the call for Q&A. If you are unable to attend, registering will still allow you to view the recordings.
Join us for three 60-minute training sessions for QIAGEN Ingenuity Pathway Analysis (IPA).
In this training, you’ll learn how to:
Tuesday July 9 – Session#1: 1pm ET/10am PT
Wednesday July 10 – Session#2: 1pm ET/10am PT
Thursday July 11 – Session#3: 1pm ET/10am PT
If you are able to attend, then we will have additional scientists on the call for Q&A. If you are unable to attend, registering will still allow you to view the recordings.
Join us for three 60-minute training sessions for QIAGEN Ingenuity Pathway Analysis (IPA).
In this training, you’ll learn how to:
Tuesday July 9 – Session#1: 1pm ET/10am PT
Wednesday July 10 – Session#2: 1pm ET/10am PT
Thursday July 11 – Session#3: 1pm ET/10am PT
If you are able to attend, then we will have additional scientists on the call for Q&A. If you are unable to attend, registering will still allow you to view the recordings.
Next-generation sequencing (NGS) oncology panels are becoming integral in hospital and academic settings to guide patient treatment and enrollment in clinical trials. But as NGS is increasingly used in precision oncology and an unprecedented amount of genomic information is uncovered, there is an industry-wide issue of standardization: a high degree of variability in variant interpretation currently exists across laboratories.
Inconsistencies in variant interpretation among laboratories can create confusion for clinicians and their patients, leaving them unable to confidently use genetic information to manage healthcare decisions. In this live panel discussion, experts in NGS testing and clinical informatics explore the issues surrounding the standardization of variant interpretation and consider how interpretation guidelines and clinical decision support (CDS) software can help to mitigate variability between laboratories.
Topics of discussion will include:
• Findings of a peer-reviewed study comparing the accuracy and consistency of variant assessments from commercial CDS software to the internal variant interpretation methods of eight laboratories.
• Causes of inter-laboratory inconsistency in variant interpretation and how to mitigate variability.
• The benefits of having an up-to-date, high-quality knowledge base to produce consistent, evidence-based clinical classifications.