

















The issue described here affects the Heat Map produced by the tool Create Heat Map for RNA-Seq when one or more layers of metadata are applied.
In affected software, heat map metadata layers are colored according to the order in which the inputs were selected, such that the left-most column receives the metadata coloring of the first input, the second column receives the metadata coloring of the second input and so on. This is incorrect because the hierarchical clustering of samples will typically re-order the samples.
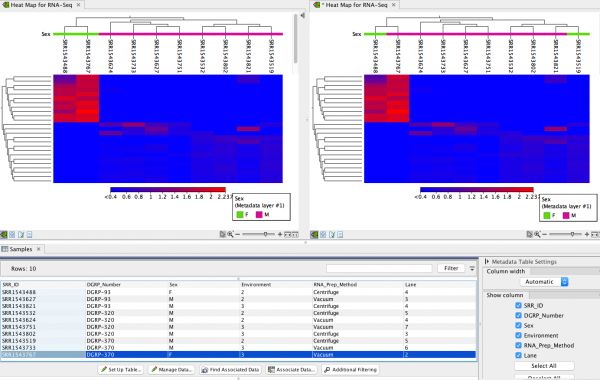
The issue is illustrated by the screenshot below. In the screenshot, the sample SRR1543767 is female (see the highlighted row in the metadata table). This is correctly shown in the Heat Map at the top left. In a Heat Map produced by affected software, shown at the top right, the sample is annotated as being male.
We recommend deleting Heat Maps produced by Create Heat Map for RNA-Seq using affected software versions and re-running these analyses with a newer version of the software.
To determine if a Heat Map was produced by the affected tool, check the software version number and tool name in the Heat Map's History view.
This issue affects heatmaps generated by Create Heat Map for RNA-Seq, whether run independently, or included in workflows, including workflows distributed by plugins, when using the following software:
This includes the following workflows delivered by QIAGEN plugins, if run on the affected software listed above:
This issue was fixed in CLC Genomics Workbench 21.0.3 and CLC Genomics Server 21.0.3.
Note: Only results of Create Heat Map for RNA-Seq are affected. Heat Maps generated by other tools are not affected.