

















The issue described here affects the tool Differential Expression in Two Groups
In affected software, normalization always proceeds using Normalization method = TMM, even when Normalization method = Housekeeping genes has been selected as in the screenshot below.
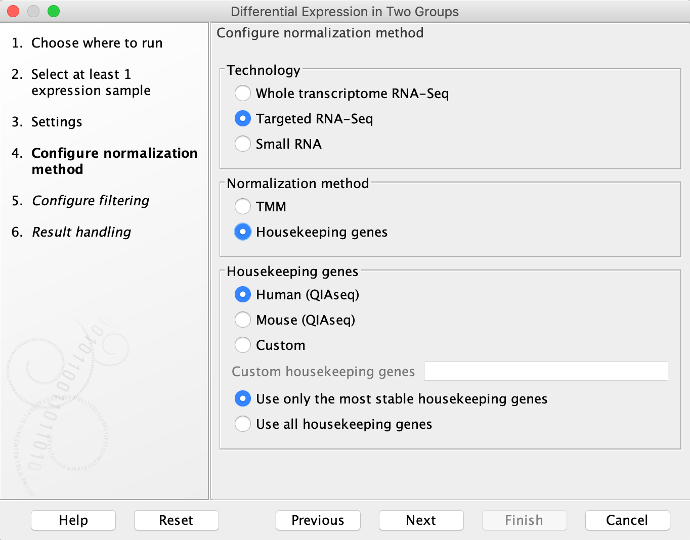
The choice of normalization method affects the calculated fold changes and p-values. In many cases, the two normalization methods will give consistent results, such that there is a very large overlap in the number of differentially expressed genes (defined by a combination of fold change and p-value cut-off).
However, results can differ substantially when the assumptions of either normalization method are violated:
It is recommended to re-run affected data with the tool Differential Expression for RNA-Seq, which is not affected by this issue. Old and new results can be compared using Create Venn Diagram for RNA-Seq.
This issue affects:
This issue was addressed in version 21.0.5 of the software.