

















Multiple issues have been identified with the Download Pathogen Database tool that result in only a subset of the intended data being downloaded, and in the data that is downloaded coming from outdated sources.
This issue affects all searches where “NCBI Pathogen Detection” has been selected.
Depending on the choice made when downloading a pathogen reference database:
Results of downstream analyses making use of affected reference data sets should be considered critically, as they will be based on a partial, possibly outdated, dataset. For example, the use of incomplete genomic assemblies could lead to incorrect strain identification and recent outbreak data may be missing completely.
For retrieving genomic data, we recommend using the Download Custom Microbial Reference Database tool, which is not affected by this issue.
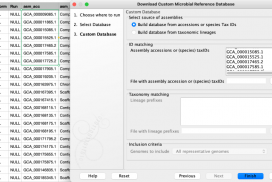
The data downloaded can now be annotated using the Create Annotated Sequence List tool by matching on the “Assembly ID” column .
The metadata file from the NCBI can be used for this, by renaming it so the suffix is .txt. E.g. for the file mentioned above, the name should be changed to “PDG000000003.1542.metadata.txt”.
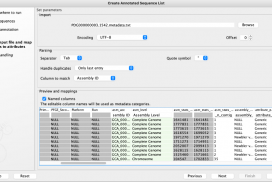
Note that renaming the headers will make the results more readable, e.g. “asm_level” to “Assembly Level”.
This issue was addressed in MGM 21.1.1