



















In the rapidly evolving field of microRNA research, which still relies heavily on a variety of measurement techniques and prediction algorithms for target identification, a common challenge is identifying the most biologically relevant targets. IPA’s new microRNA Target Filter functionality enables prioritization of experimentally validated and predicted mRNA targets.
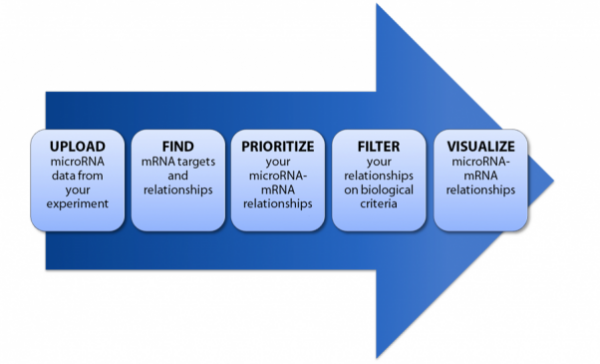
microRNA workflow in IPA
Upload, analyze, prioritize, filter, and visualize microRNA-mRNA data and relationships, all within a single tool. You can also leverage microRNA and mRNA data in combination with other types of ‘omics and high-throughput data for a fully integrated biological analysis. Using IPA in your microRNA research provides you with enhanced knowledge about subcellular location, functional gene family, association with drugs and pathways, and more.
Highlights
The microRNA Target Filter in IPA provides insights into the biological effects of microRNAs, using experimentally validated interactions from TarBase and miRecords, as well as predicted microRNA-mRNA interactions from TargetScan. Additionally, IPA includes a large number of microRNA-related findings from the peer-reviewed literature.
IPA reduces the number of steps it takes to confidently, quickly, and easily identify mRNA targets by letting you examine microRNA-mRNA pairings, explore the related biological context, and filter based on relevant biological information as well as the expression information. The ability to leverage biological context is key to overcoming the inherent complexity in current microRNA data analysis. Using IPA, you can easily and consistently answer questions like:
Capabilities
MicroRNA content:
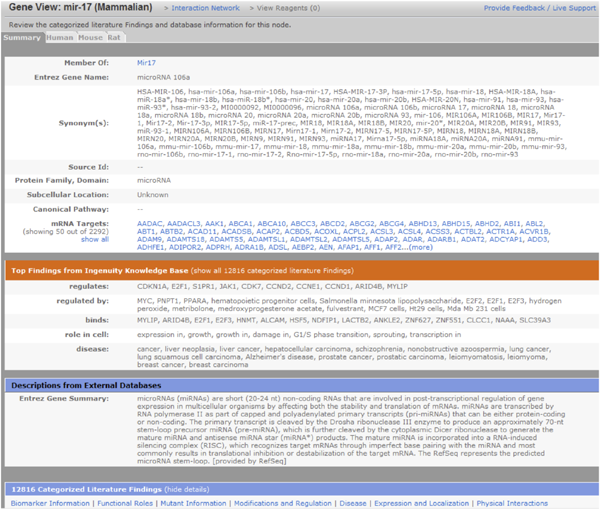
Search for mRNA targets, disease states, synonyms, regulators, biomarkers, expression and more in IPA using microRNA content in the Ingenuity Knowledge Base. (Example above is a Gene View for Entrez Gene miR-106, which is within family miR-17. MicroRNA IDs are from miRBase and predicted relationships are from TargetScan.)
MicroRNA Target Filter
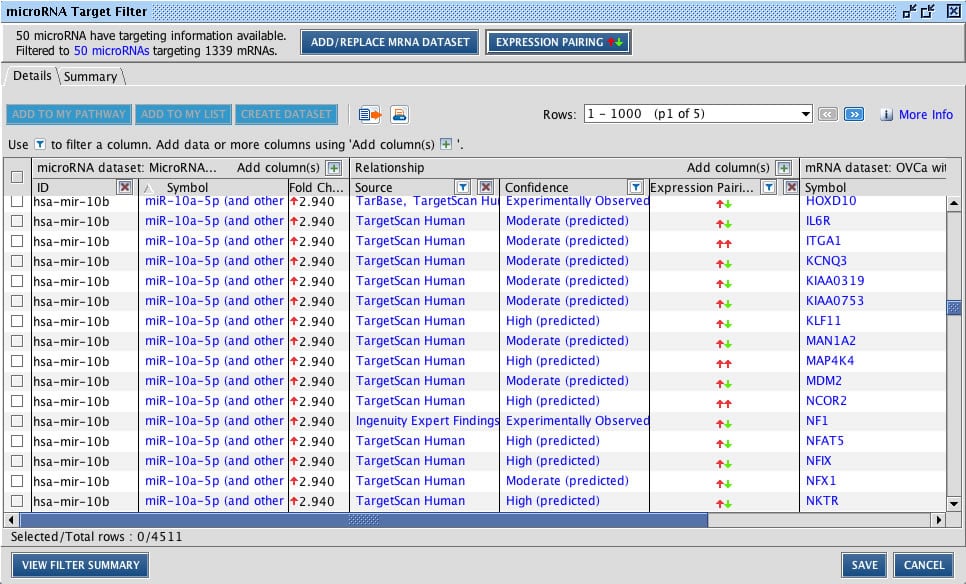
No other tool lets you identify, filter, and prioritize microRNA-mRNA relationships in one easy step.
Above, the MicroRNA Target Filter ready to prioritize relationships based upon molecular type or pathway.
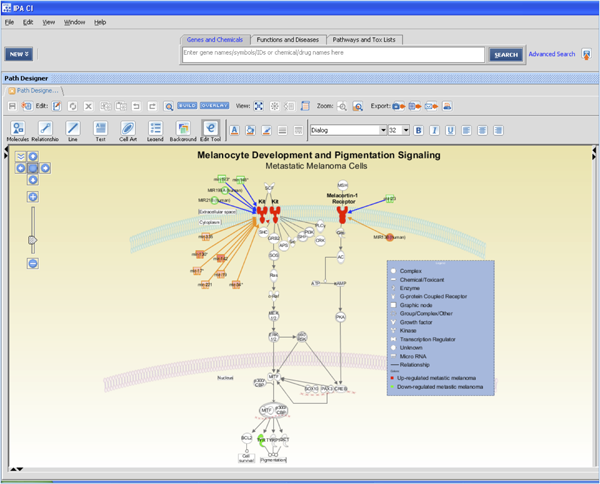
Once key relationships are identified, they can be exported to a pathway for visualization and exploration. In this case, from over 13,000 potential targets, IPA helped identify that just two are the top proteins – KIT and MC1R – driving melanocyte development and pigmentation signaling. They are targeted by about 10 different microRNAs, 5 of which have inverse expression patterns in metastatic melanoma.