


















In the high-stakes field of pharmacogenomics, precision and accuracy are non-negotiable. Evidence-based databases in pharmacogenomics are indispensable tools that enable labs to interpret genetic variants confidently and accurately. But how can labs integrate these resources effectively to deliver reliable and timely annotations?
As pharmacogenomics (PGx) becomes an essential tool in precision medicine, the accuracy, consistency, and relevance of genetic interpretations are paramount. Clinicians and laboratory scientists depend on insights from genetic variants to guide drug therapy decisions, making it critical that these interpretations are grounded in high-quality, evidence-based data. Trusted databases, such as PharmVar, CPIC, and PubMed, serve as vital resources for interpreting the impact of genetic variations on drug response. However, leveraging these databases effectively—and avoiding outdated or low-quality sources—is essential to ensuring the most accurate, relevant, and timely care.
Pharmacogenomic testing is designed to reveal how an individual’s genetic makeup can influence their response to medications, offering personalized therapeutic options that can enhance efficacy and minimize adverse effects. But the success of this approach hinges on the correct interpretation of genetic variants. The sheer complexity of the human genome, combined with the variability in drug-gene interactions, makes it crucial for clinicians and scientists to rely on validated, up-to-date resources. Misinformed or incomplete interpretations can lead to suboptimal drug choices, ineffective dosing, or serious safety concerns.
Accurate PGx interpretation requires more than raw genetic data—it demands a multidisciplinary approach that integrates trusted and continually updated resources. Below are five PGx sources that provide essential insights that, when combined, offer a comprehensive framework for understanding gene-drug interactions.
The U.S. Food and Drug Administration (FDA) offers crucial information on how genetic variations affect drug efficacy, dosing, and safety through its Table of Pharmacogenomic Biomarkers in Drug Labeling. This resource identifies drugs with pharmacogenomic information in their labels, signaling when genetic testing might be relevant for prescribing decisions. Labs and pharmaceutical companies can use this FDA-approved data to determine whether a pharmacogenomic analysis should influence drug choice or dosage recommendations.
For example, labs can use FDA data to look for biomarker-drug associations. The FDA table identifies gene-drug interactions, specifying genetic markers that impact drug metabolism or response. Drugs like warfarin (with CYP2C9 and VKORC1 variants) and clopidogrel (with CYP2C19) are directly linked to genetic markers that affect patient outcomes.
The Clinical Pharmacogenetics Implementation Consortium (CPIC) is an essential tool for translating genetic data into therapeutic insights. The CPIC guidelines provide scientists and clinicians with peer-reviewed, evidence-based data on how specific genetic variants should inform drug prescribing. Unlike many other resources, CPIC focuses on "how to use" genetic test results rather than "whether to test," thereby empowering labs to make confident, well-informed decisions. The continuously updated nature of CPIC guidelines ensures that they reflect the latest research and clinical practices, helping labs avoid outdated interpretations that could compromise outcomes.
For scientists tasked with generating pharmacogenomic insights, referencing CPIC ensures that their interpretations align with the most current, validated data. This is particularly important when dealing with complex metabolizer phenotypes, such as poor or ultra-rapid metabolizers, where small interpretation errors can lead to significant consequences.
The Dutch Pharmacogenetics Working Group (DPWG) provides another vital resource for interpreting PGx data, offering global perspectives that complement CPIC’s guidelines. The DPWG guidelines, developed with rigorous scientific evidence, provide insights on adjusting drug regimens based on genetic test results. They are particularly helpful when dealing with drugs commonly used in European populations or in international settings.
PharmVar (Pharmacogene Variation Consortium) plays a key role in standardizing the nomenclature of pharmacogenomic variants, especially within critical genes like CYP2D6, CYP2C9, and CYP2C19, which influence the metabolism of many drugs. PharmVar curates alleles and haplotypes, ensuring that scientists and clinicians have consistent and accurate variant information for pharmacogenomic interpretation.
Labs can use PharmVar data to access standardized allele designations. PharmVar’s consistent allele nomenclature allows for accurate interpretation of genetic variants. This is particularly important when dealing with genes with complex variant structures, like CYP2D6, which has multiple alleles and copy number variations.
PubMed, maintained by the National Library of Medicine, is an indispensable database for accessing peer-reviewed scientific literature. Labs interpreting pharmacogenomic markers can use PubMed to stay up-to-date on the latest research findings, clinical trials, and pharmacogenomic studies that inform gene-drug interactions.
Labs can use PubMed to search for specific gene-drug pairs or PGx markers and find studies that validate or expand upon the data found in FDA labeling, CPIC, or DPWG guidelines. In addition, PubMed enables the evaluation of emerging evidence. PGx is a rapidly evolving field. PubMed allows labs to keep abreast of emerging research on rare variants, newly discovered gene-drug interactions, or novel therapies with PGx implications.
Currently, most laboratories engaged in PGx interpretation must consult a variety of resources, databases, and guidelines to comprehensively annotate test results. While these resources are invaluable, they are often siloed, leading to challenges in consistency and efficiency. Laboratories must manually piece together information from multiple sources, which can be fragmented or even outdated. On average, manual PGx annotation can take hours—even days—making it difficult to deliver reliable and timely annotations Furthermore, discrepancies between databases or inconsistent variant nomenclature can further complicate clinical decision-making.
This fragmented approach underscores the need for a single, authoritative knowledgebase that centralizes all relevant PGx resources. A unified platform that integrates expert-curated data from multiple sources, continuously updated for accuracy and relevance, would streamline the annotation process. Such a resource would eliminate redundancy, improve consistency, and provide labs with a reliable foundation for interpreting PGx test results. This would not only enhance efficiency but also increase the utility of pharmacogenomic testing, ensuring that care is informed by the most up-to-date and accurate genetic insights.
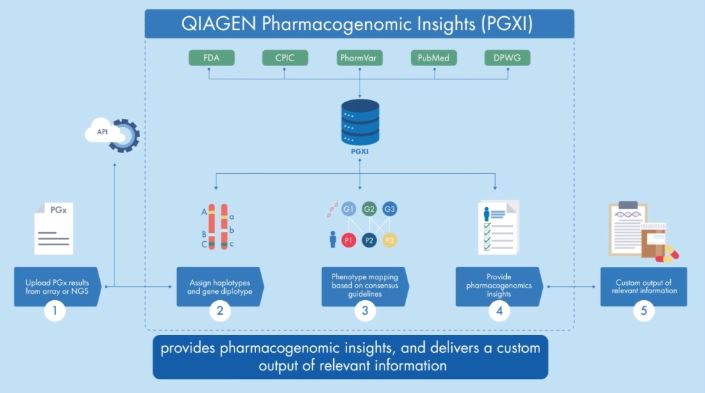
QIAGEN Pharmacogenomic Insights (PGXI) is a new knowledgebase designed to help scientific and translational researchers better understand how an individual's genetic make-up influences their response to medications. Providing a streamlined and automated approach to how labs can efficiently and confidently produce evidence-backed insights for pharmacogenomic markers, PGXI enables faster, more precise drug development and delivery.
PGXI is a new knowledgebase solution within PGx built upon a legacy platform used to deliver more than 1.6 million PGx annotations for more than 250 clinical research laboratories. The knowledgebase contains data from all established pharmacogenomic information sources, including the FDA, CPIC, DPWG, PharmVar, and curated articles from the biomedical literature database PubMed, to provide accurate, up-to-date evidence in one centralized location. Users can submit pharmacogenomic test results to PGXI and rapidly query and identify relevant gene-drug associations and conditions linked to specific genotypes.
PGXI is transforming precision medicine by dramatically speeding up the process of analyzing how a person’s genes affect their response to medications. Within minutes, users can retrieve a custom output of relevant information, including:
For labs and pharmaceutical companies needing an easier and faster way to translate complex pharmacogenomic data into evidence-backed insights to inform drug delivery and development, QIAGEN PGXI provides a better a way forward.
→ Learn more about PGXI and request a free trial here.

Learn how to use PGXI to rapidly and confidently translate complex PGx data into evidence-backed therapeutic insights.

Learn more about PGXI, including how the knowledgebase can integrate into new or existing systems.
Gain a better understanding of how your lab can use PGXI to rapidly translate PGx markers into therapeutic insights.