

















A recent paper presents the ChIP-seq analysis tool available in CLC Genomics Workbench and CLC Genomics Server (version 7.5 and later).
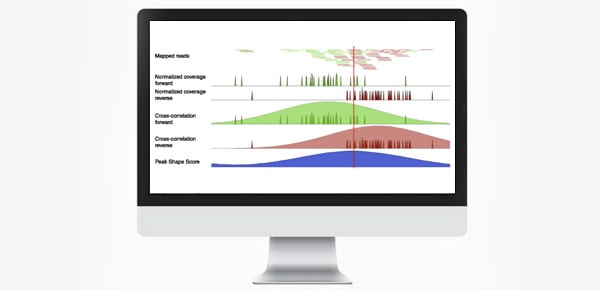
Some of the main requirements for the development of our peak calling toolset were generality, specificity, robustness, and simplicity. This resulted in the CLC shape-based peak caller which implements all the steps of signal detection, quality control, normalisation, discovering obvious peaks, learning the peak shape, peak shape score, and peak detection in a single, easy-to-use algorithm. The algorithm delivers a QC report containing metrics about the quality of the ChIP-seq experiment, a peak shape score value for every genomic position, and a list of all called peaks.
We have showed from a performance evaluation that the CLC shape-based peak caller ranks well among popular state-of-the-art peak callers while requiring a minimum of intervention and parameterization from the user.
Read the whole article: Identifying peaks in *-seq data using shape information (Lappe and Strino, 2016)
Get more on our solution for epigenomics analysis