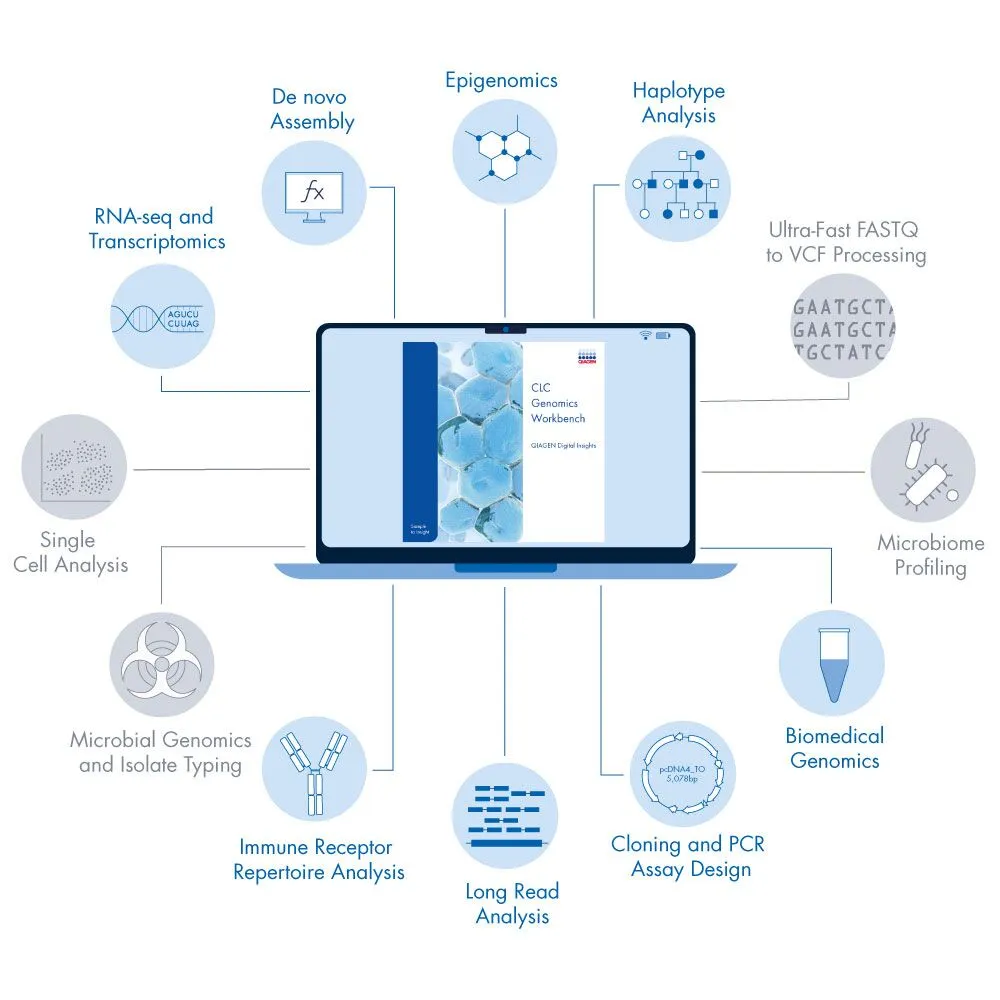
QIAGEN CLC Genomics Workbench
A complete toolkit for genomics, transcriptomics, epigenomics, and metagenomics analysis in one program.

Home > Bioinformatics Products Overview > Bioinformatics Tools and Applications > Genomics Data Analysis Software > QIAGEN CLC Genomics Workbench
Sequence analysis software for any species, any platform, any workflow
QIAGEN CLC Genomics Workbench is a powerful solution that works for everyone, no matter the workflow. Cutting-edge technology and unique features and algorithms widely used by scientific leaders in industry and academia make it easy to overcome challenges associated with data analysis.
Scale your research using QIAGEN CLC Genomics Workbench 23
Now featuring template workflows, import of RNA-seq expression data in matrix format, fast long-read assembler for large eukaryotic genomes, de novo prediction of eukaryotic genes using GeneMark-ES and methylation profiling.
Read about the latest improvements.
Need a full set of features, including microbial and metagenomics, single cell analysis or processing of small genomes? Discover our full-featured solution, QIAGEN CLC Genomics Workbench Premium.
Complete toolkit for genomics, transcriptomics, epigenomics, and metagenomics in one program
Comprehensive NGS data analysis
User-friendly bioinformatics software solutions allow for comprehensive analysis of your NGS data, including de novo assembly of whole genomes and transcriptomes, resequencing analysis (WGS, WES and targeted panel support), variant calling, RNA-seq, ChIP-seq and DNA methylation (bisulfite sequencing analysis).
Analyze your RNA-seq and small RNA (miRNA, lncRNA) data with easy-to-use transcriptomics workflows for differential expression analysis at gene and transcript levels.
Supported NGS platforms include Illumina, IonTorrent, Oxford Nanopore and PacBio.
Biomedical genomics analysis
Biomedical genomics analysis and panel data analysis functionality is delivered through the QIAGEN CLC Genomics Workbench and the free plugin, Biomedical Genomics Analysis. This plugin provides tools and workflows for NGS panel data analysis, including WES, WGS and RNA-seq, as well as SARS-CoV-2 panel analysis workflows for QIAseq and Ion AmpliSeq.
Template workflows
Biomedical workflows with complete reference sets for human, mouse and rat genomics include hereditary disease workflows (trio analysis, family-of-four) and oncology somatic mutation detection workflows, and can be used on FFPE or liquid biopsy samples (single sample or tumor-normal matched samples). These workflows enable sensitive detection of SNPs, MNVs, InDels, tandem repeats, structural variants, fusion genes, CNVs and loss of heterozygosity (LoH). Annotation with conservation scores and filtering steps on dbSNP and ClinVar are included.
Extend the toolbox with plugins
Multiple plugins are available for the QIAGEN CLC Genomics Workbench, tailored to specific applications such as multiple sequence alignment, whole genome alignment, transcript discovery, biomedical genomics analysis, long read analysis and haplotype calling. These feature-rich extensions are seamlessly integrated into the QIAGEN CLC Genomics Workbench, and provide advanced tools and workflows to meet your specific needs for analysis.
Uncover the signals that lead to breakthrough discoveries in your biomedical research
Support for the analysis of NGS data from human and mouse PCR-based GeneRead panels or Unique Molecular Index (UMI)-based miRNA, RNA, DNA and methylation QIAseq panels is also provided by the Biomedical Genomics Analysis plugin. The tools and workflows work as well with third-party vendor NGS panels (PCR, UMI or hybrid capture target enrichment panels such as Illumina TSO500, Ion Torrent, ArcherDx, Agilent, NimbleGen or Twist panels). Read our Case Study: QIAseq Myeloid Neoplasm Panel

Download QIAGEN CLC Genomics Workbench and Enjoy a FREE full-feature trial for 14 days
QIAGEN CLC Genomics Workbench is developed for Windows, Mac and Linux. Download now to start your data analysis. To get started follow the step by step instructions in the user-friendly manual or watch the tutorials in our resources guide.
Enterprise Solutions
Enhancing your productivity is our business
QIAGEN CLC Genomics Workbench allows scaling to any size. Enterprise solutions permit entire institutions to make use of the same software.
Customized workflows to accelerate your data analysis
QIAGEN CLC Genomics Workbench is developed to support a wide range of NGS bioinformatics applications. Workflows can combine quality control steps, adapter trimming, read mapping, variant detection, and multiple filtering and annotation steps into a pipeline you can share with colleagues and execute with just one click.
New workflow elements together with enhanced metadata capabilities allow within-workflow batching, useful for e.g., iterating the quantification of many RNA-seq samples followed by statistical comparison of groups.
Let QIAGEN CLC Genomics support you in the classroom
Want to develop an NGS data analysis curriculum while advancing your own research goals? Check out the benefits of our teaching license package to QIAGEN CLC Genomics Workbench.
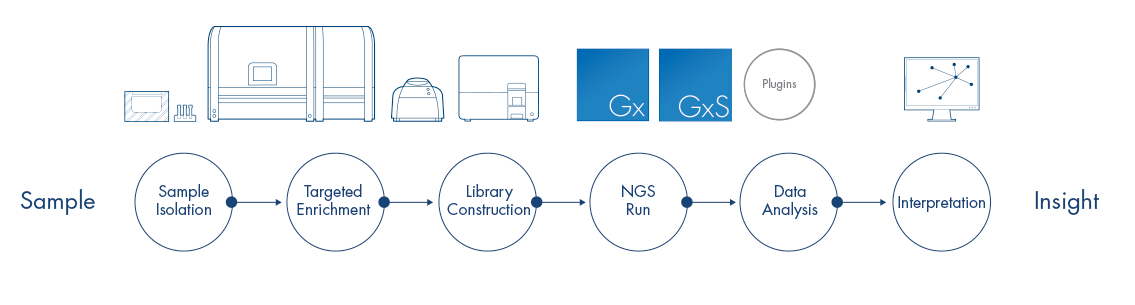
From sample to insight with QIAGEN Digital Insights
From QIAGEN CLC Genomics Workbench you have easy access to pre-formatted reference data sets for many species. You can also submit results to other solutions in the QIAGEN Digital Insights portfolio, such as Ingenuity Pathway Analysis (IPA), QIAGEN Clinical Insight Interpret (QCI) and QIAGEN Clinical Insight Interpret Translational (QCI-IT).
Download a Free Trial
QIAGEN CLC Genomics Workbench is a powerful solution that works for everyone, no matter the workflow.
Request a Quote
Need a full set of features, including microbial and metagenomics, single cell analysis or processing of small genomes?
Publications
Discovery the diversity of research for which QIAGEN CLC Genomics Workbench makes a difference.

Latest improvements
We frequently release updates and improvements such as new functionalities, bug fixes or plugins. To get a complete overview, please read the latest improvements.
System Requirements
- Windows 10, Windows 11, Windows Server 2012, Windows Server 2016, Windows Server 2019 and Windows Server 2022
- Mac: macOS 11, 12, and 13
- Linux: RHEL 7 and later, SUSE Linux Enterprise Server 12 and later. The software is expected to run without problem on other recent Linux systems, but we do not guarantee this. To use BLAST related functionality, libnsl.so.1 is required.
- 64 bit operating system
- 1 GB RAM required
- 2 GB RAM recommended
- 1024 x 768 display required
- 1600 x 1200 display recommended
System requirements for 3D viewers
- Requirements
- A graphics card capable of supporting OpenGL 2.0.
- Updated graphics drivers. Please make sure the latest driver for the graphics card is installed.
- Recommendations
- A discrete graphics card from either Nvidia or AMD. Modern integrated graphics cards (such as the Intel HD Graphics series) may also be used, but these are usually slower than the discrete cards.
3D rendering is only supported when the CLC Main Workbench is installed on the same machine the viewer is opened on: indirect rendering (such as x11 forwarding through ssh), remote desktop connection/VNC, and running in virtual machines is not supported.