Interpretation of genomic variants in tumor samples still presents a challenge in clinical settings. Variant interpretation is fragmented across disparate databases, and aggregation of information from these requires building extensive infrastructure. The Human Somatic Mutation Database (HSMD) is an easy-to-use, somatic database from QIAGEN that contains extensive genomic content relevant to solid tumors and hematological malignancies. Pulling content from over 500,000 real-world clinical oncology cases and 40+ databases contained in the QIAGEN Knowledge Base, HSMD gives genetic counselors access to over 1.7 million somatic variants characterized in over 1,400 cancer-related genes. To demonstrate the efficiency and clinical utility of HSMD, we present a use-case for using the database to assess the biological and clinical relevance of anaplastic lymphoma kinase (ALK) gene.
The ALK gene
About 3-7% of patients with non-small cell lung cancer (NSCLC) have rearrangements in the ALK gene. These genetic alterations are relatively rare compared with epidermal growth factor receptor (EGFR) or KRAS mutations (1). ALK rearrangements are often seen in people who don’t smoke and who are younger. These oncogenic mutations lead to the constitutive activation of the ALK tyrosine kinase domain, a protein that causes cancer cells to grow and spread. To date, several therapies have been developed to target ALK gene changes, known as ALK inhibitors. These include, Crizotinib (Xalkori), Ceritinib (Zykadia), Alectinib (Alecensa), Brigatinib (Alunbrig), and Lorlatinib (Lorbrena).
Therefore, clinical diagnostic labs that perform somatic genomic testing need to be able to define the precise biological function and clinical actionability of ALK mutations. However, given their relatively rarity, ALK mutations require thorough assessment and curation to accurately interpret their clinical significance and recommend effective treatment strategies.
Using HSMD to assess the biological and clinical significance of ALK mutations
HSMD is a web-based database that provides deep insight into small variants, such as SNVs, indels, frameshifts, fusions and copy number variants that have been clinically observed or curated from scientific literature to help users better understand and define precise function and actionability. Unlike other somatic databases, such as ClinVar and Genomenon that use crowdsourcing or human-absent machine learning, HSMD uses augmented molecular intelligence—the combination of machine learning and human curation—to curate each somatic variant. The result is higher quality data that is up-to-date, consistent, comprehensive, and accurate. This focus on human effort, review and certification is critical. It means clinical diagnostic labs can trust the data in HSMD and proceed with confidence to interpret and report somatic tests.
Here, we present a step-by-step use-case of how your clinical diagnostic lab can use HSMD to search and explore mutational characteristics across the ALK gene, synthesize key findings from drug labels, clinical trials, and professional guidelines, and receive detailed annotations for each observed variant.
Searching for the ALK gene in HSMD
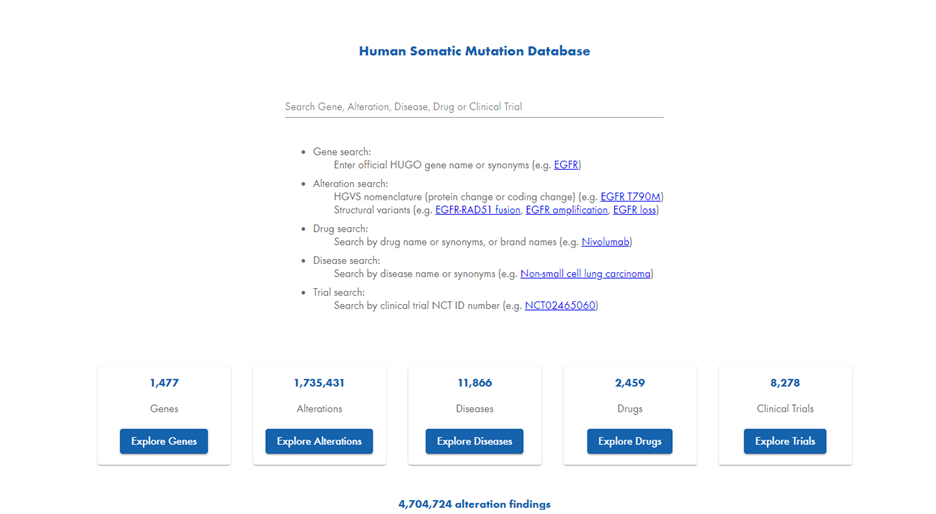
HSMD is a web-based application. When you open the interface, the homepage provides five search options. Users can search by Gene, Alteration, Disease, Drug, or Clinical Trial (Figure 1).
Search by gene: When you search by gene in HSMD, you access the total number of genes listed in the database. You can then further narrow your search by viewing data from the QIAGEN Knowledge Base or focusing exclusively on clinically observed variants.
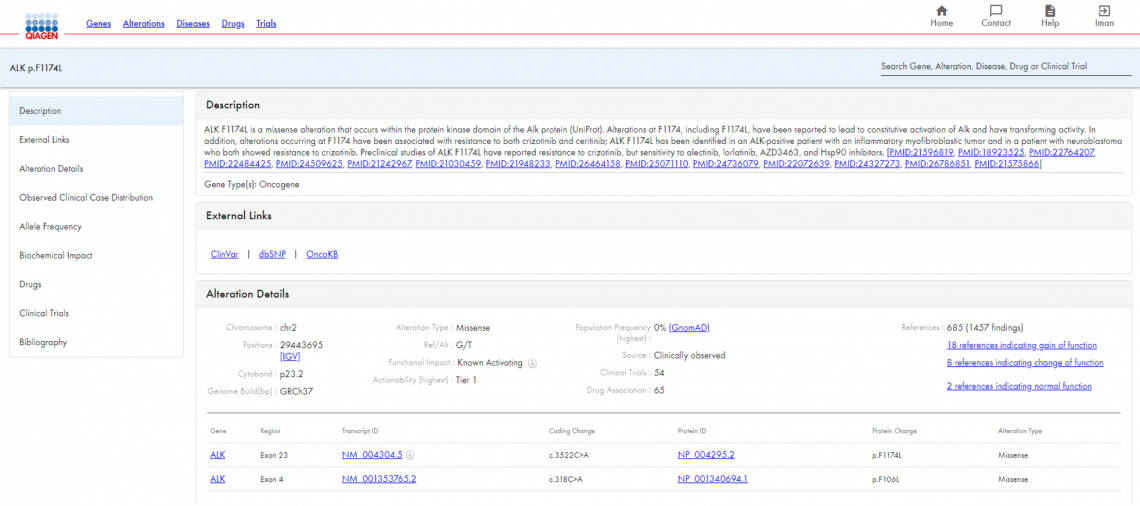
When you search for ALK p.F1174L in HSMD, you receive a description of the alteration with links to relevant literature, external links to the alteration in ClinVar, dbSNP, and OncoKB, and a summary of alteration details, including chromosome position, alteration type, functional impact, and population frequency (Figure 2).
View observed clinical case distribution of ALK in HSMD
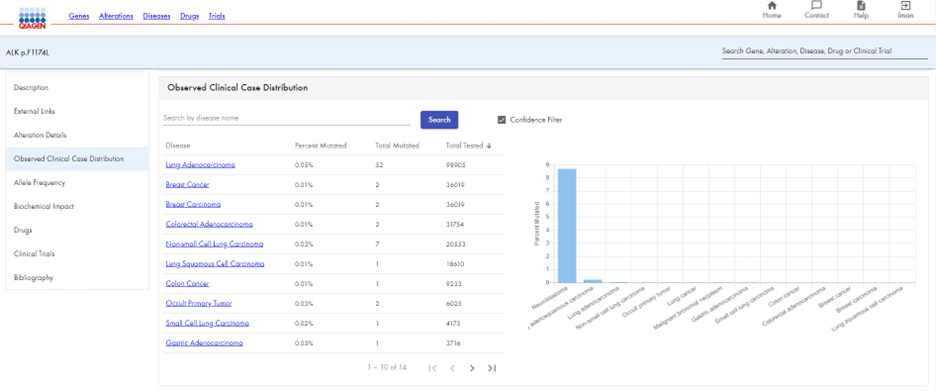
Once you narrow your focus to searching for only clinically observed variants, you receive a table and distribution graph that shows the number of observed clinical cases for a particular gene across all cancer types.
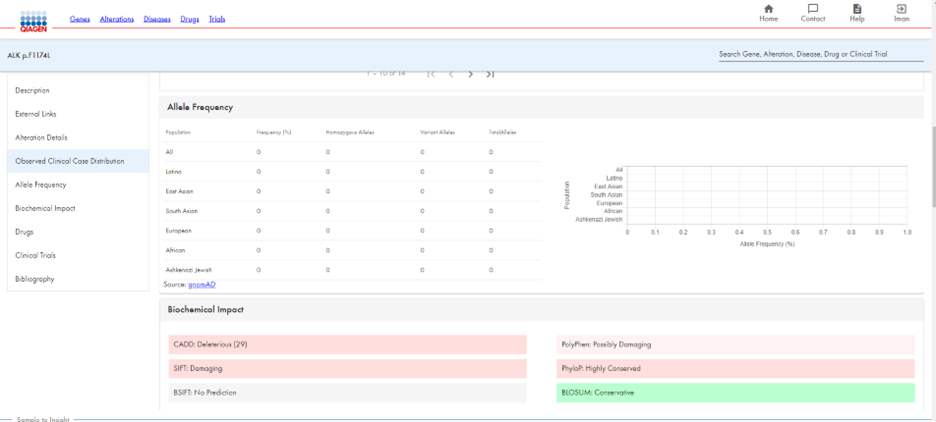
Continuing the example using ALK p.F1174L, you can clearly see the alteration’s distribution by disease through a table and graph. As you scroll down, you receive information on the gene’s frequency in different populations, as well as its biological impact (Figure 3). You can also view the allele frequency and biological impact. (Figure 4).
Click to enlarge the images.
View relevant drugs, clinical trials and bibliography for ALK in HSMD
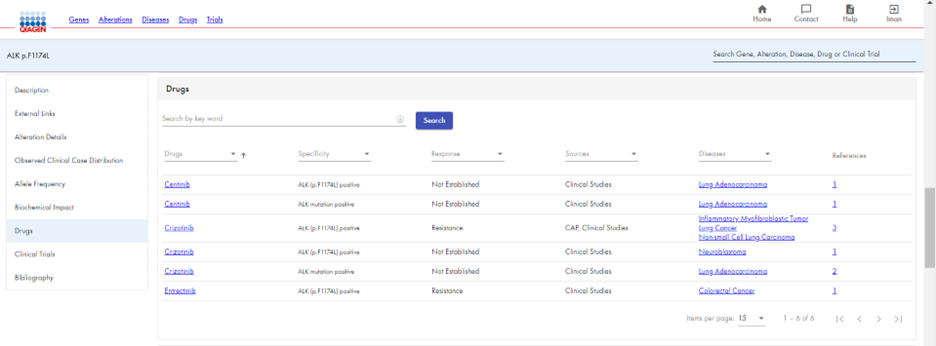
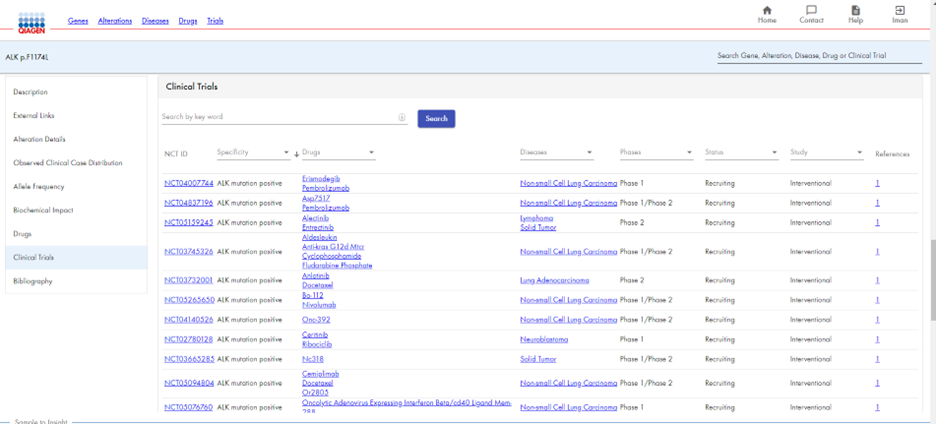
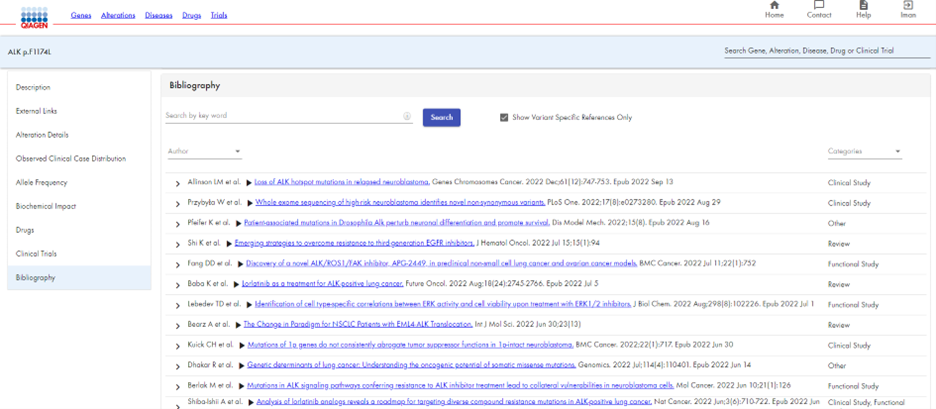
HSMD enables you to view relevant drugs and clinical trials for a ALK p.F1174L. You receive a list of drugs approved for use with ALK p.F1174L (Figure 5a), as well as recruiting clinical trials for that alteration (Figure 5b). In addition, HSMD provides an extensive bibliography for ALK p.F1174L, with clickable links to each article (Figure 6).
Click to enlarge the images.
Simplify your somatic variant interpretation
As demonstrated by the use-case, incorporating HSMD into your somatic variant interpretation workflow enables you to easily search and explore mutational characteristics across genes, synthesize key findings from drug labels, clinical trials, and professional guidelines, and receive detailed annotations for each observed variant. With HSMD, you can efficiently ask and answer the following key questions:
- Is this patient eligible for targeted treatment based on the molecular profile?
- Is there a clinical trial that could be considered for the patient?
- Where is the clinical trial? Is there an off-label therapy?
Want to try HSMD for free?
HSMD is a one-stop shop for all the content a clinical diagnostic lab needs to assess the biological relevance and clinical actionable of somatic variants. QIAGEN Digital Insights offers free, no-obligation trials of HSMD. You can see what kind of content the database offers, explore the search functionality, and determine if this database can save you time and money.
Need a FASTQ to final report solution for oncology NGS testing?
References
- Cha, Y.J., Kim, H.R. & Shim, H.S. Clinical outcomes in ALK-rearranged lung adenocarcinomas according to ALK fusion variants. J Transl Med 14, 296 (2016). https://doi.org/10.1186/s12967-016-1061-z