
Variants that span a target region are not called
Issue description
When calling variants with the Basic Variant Detection, Low Frequency Variant Detection, or Fixed Ploidy Variant Detection tool and using the “Restrict calling to target regions” option, variants that span the target region, that is, overlap with and are longer than, the target region, are not detected.
This issue would primarily affect the detection of deletions. An example where this might arise would be where a deletion occurred at the DNA level, but the target was defined based on an exon. If the deletion was longer than the exon, it would not be detected.
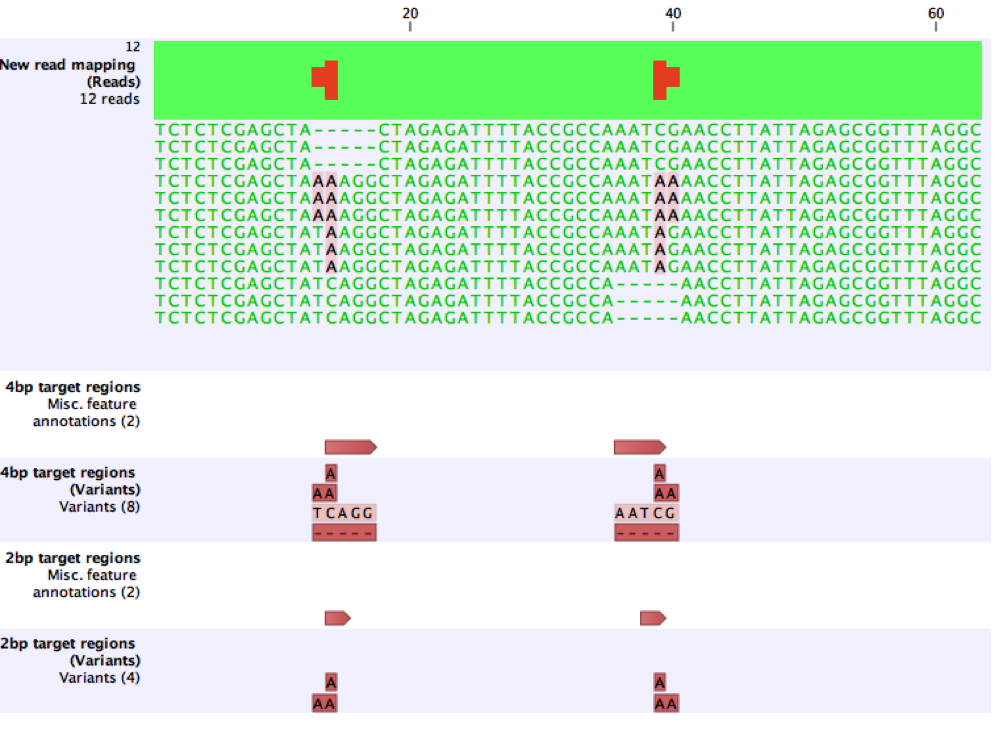
The effect of this is illustrated on a small synthetic example. Here, when 4bp target regions are provided, variants are called as expected. When the target regions are reduced to 2bp, the deletions are no longer called because they span the target regions, but other variants are still called as expected.
Affected software
- CLC Genomics Workbench 6.5 to 12.0.2
- CLC Genomics Server 5.5 to 11.0.2
- All versions of Biomedical Genomics Workbench
From CLC Genomics Workbench 12.0.3 and CLC Genomics Server 11.0.3, variants extending up to 50 nucleotides beyond either end of a target region are now reported in full, while variants extending even further will include only the first 50 nucleotides beyond the target region. Insertions at the right hand border of a target region are now considered to be a variant within the target region.