Home > Research and Discovery > Gene Expression > Scientific spotlight
Paper: Simulation-based comprehensive benchmarking of RNA-seq aligners
Giacomo Baruzzo, et al., performed a comprehensive benchmarking of 14 common splice-aware aligners for base, read, and exon junction-level accuracy and compared default with optimized parameters. The study found CLC Genomics Workbench to be one of the most reliable general-purpose aligners.
Baruzzo G, Hayer KE, Kim EJ, Di Camillo B, FitzGerald GA, Grant GR. Simulation-based comprehensive benchmarking of RNA-seq aligners. Nature methods. 2017;14(2):135-139. doi:10.1038/nmeth.4106.
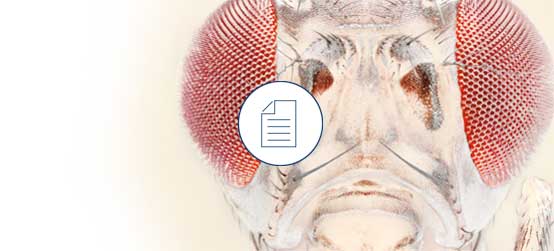
Paper: Comprehensive transcriptome analysis of early male and female Bactrocera jarvisi embryos
Jennifer L. Morrow, et al. use CLC Genomics Workbench to analyze the transcriptome of the fruit fly Bactrocera jarvisi, a horticultural pest, generating new insights into candidate genes for transgenic pest-management strategies.
Jennifer L Morrow, Markus Riegler, A Stuart Gilchrist, Deborah CA Shearman and Marianne Frommer (2014). Comprehensive transcriptome analysis of early male and female Bactrocera jarvisi embryos. BMC Genetics, 15(Suppl 2), S7. doi:10.1186/1471-2156-15-S2-S7
Application note: How to use CLC RNA-Seq analysis tools with a de novo assembled transcriptome as reference
CLC bio’s RNA-Seq tools allow for expression analysis in organisms without previously sequenced genomes. This is especially important in plant and animal genome research where the majority of species do not have reference sequences available. The CLC bio de novo assembler and RNA-Seq tools have been used for years by many plant and animal researchers. This document provides basic guidelines for RNA-Seq analysis using de novo assembled transcripts as reference.
Paper: Investigation of the retinal transcriptome and molecular pathways using RNA-Seq technology may open new avenues to treatment strategies
Glaucoma is a leading cause of blindness worldwide and thus an important disease to understand. The disease is characterized by retinal ganglion cell (RGC) death caused by axonal injury and in glaucoma patients the number of RGCs decreases due to axonal degeneration, resulting in visual dysfunction.
RNA-Seq was used to investigate the entire retinal transcriptome profile in the early stages of post-axonal injury. Sequencing data was imported to CLC Genomics Workbench for RNA-Seq, gene expression and statistical analysis.
Masayuki Yasuda, Yuji Tanaka, Morin Ryu, Satoru Tsuda, Toru Nakazawa RNA Sequence Reveals Mouse Retinal Transcriptome Changes Early after Axonal Injury PLoS ONE 9(3): e93258. doi:10.1371/journal.pone.0093258