
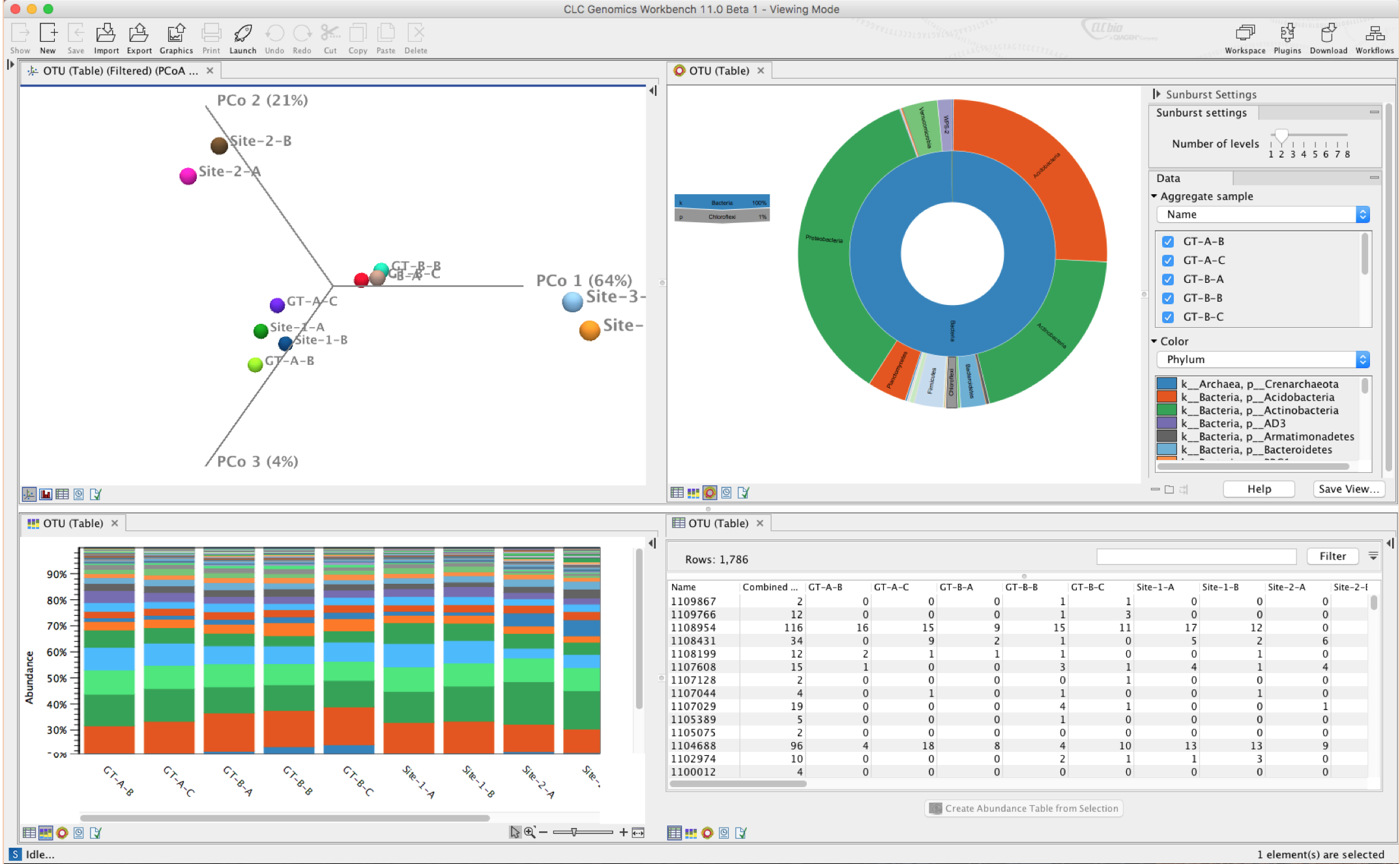
CLC Genomics Workbench 11
Share insights with the new Viewing Mode
Do you need a free genome browser for data sharing and collaboration? Look no further – Biomedical Genomics Workbench and CLC Genomics Workbench can now be used as free genome browsers to share, view, and explore NGS analysis results. Enjoy support for a wide range of open and proprietary file formats. No License required!
- All bioinformatics file importers and exporters have been enabled in Viewing Mode for sharing and viewing of a wide range of file formats.
- To start using the new Viewing Mode install the workbench of your choice. When the workbench asks for a license, simply choose to run in Viewing mode.

Share module network licenses more freely
Good news for site license owners: We have improved sharing of Modules for CLC Workbenches. Administrating network licenses and sharing module licenses with colleagues is now more flexible. • Workbenches now only request a Module license from the CLC License Server when a tool from a module is actually launched. • Workbenches return the Module license 4 hours after the last time a tool was launched, even if the user has not closed the Workbench. • As before, all acquired Module licenses are released to the CLC License Server when the Workbench is closed. • Colleagues may install a Module and explore the results without needing a license at all as long as no tool is launched. • Stay tuned for more: we are working on an improved user interface for the CLC License Server.
Avoid analysis error through improved adapter trimming
New: Read-through adapter trimming
With growing read length, NGS reads more and more often also include partial downstream adapter sequences at the read end. Such often very short read-through adapter fragments are hard to detect by conventional methods, but if untrimmed can introduce errors to variant detection or de novo assembly. Most adapter trimming tools available to researchers today fail to detect these artefacts. We have skimmed published datasets and found that on average~6% of the reads do still include partial read-through adapters.
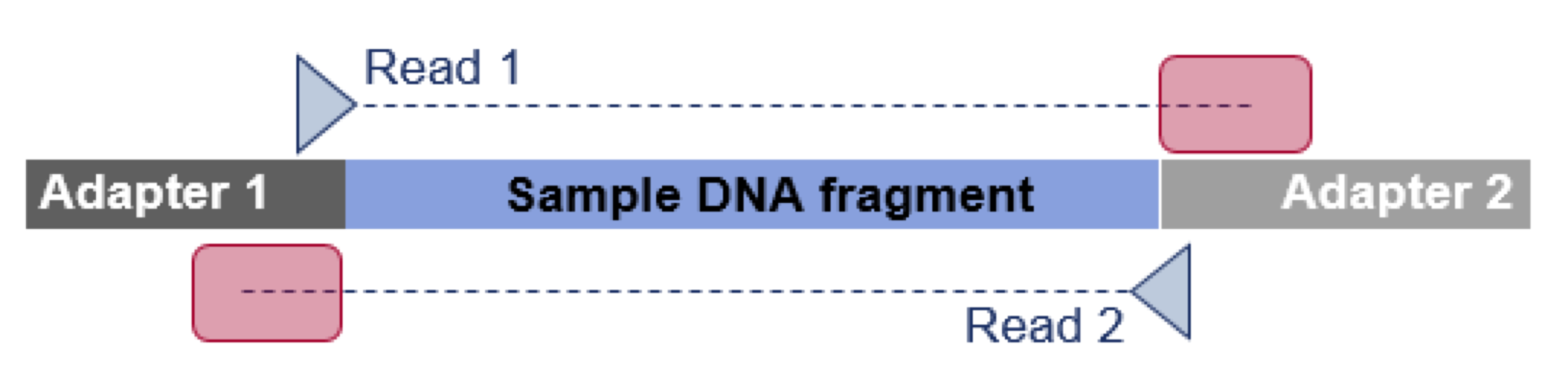
In this example, the sequenced NGS read is longer than the DNA fragment. As a consequence, the reads will extend into the downstream “read-through” adapter (see red box).
To reduce the chance for error we are introducing effective read-through adapter trimming for paired read data.
- Automatic detection and trimming of read-through adapters. No manual configuration required.
- Since automatic detection works by comparing first and the second read of a Paired-end pair, this option is only available for Paired-end reads. Automatic adapter-trimming will only detect read-throughs, and can be combined with general adapter trimming for the highest accuracy of adapter trimming.
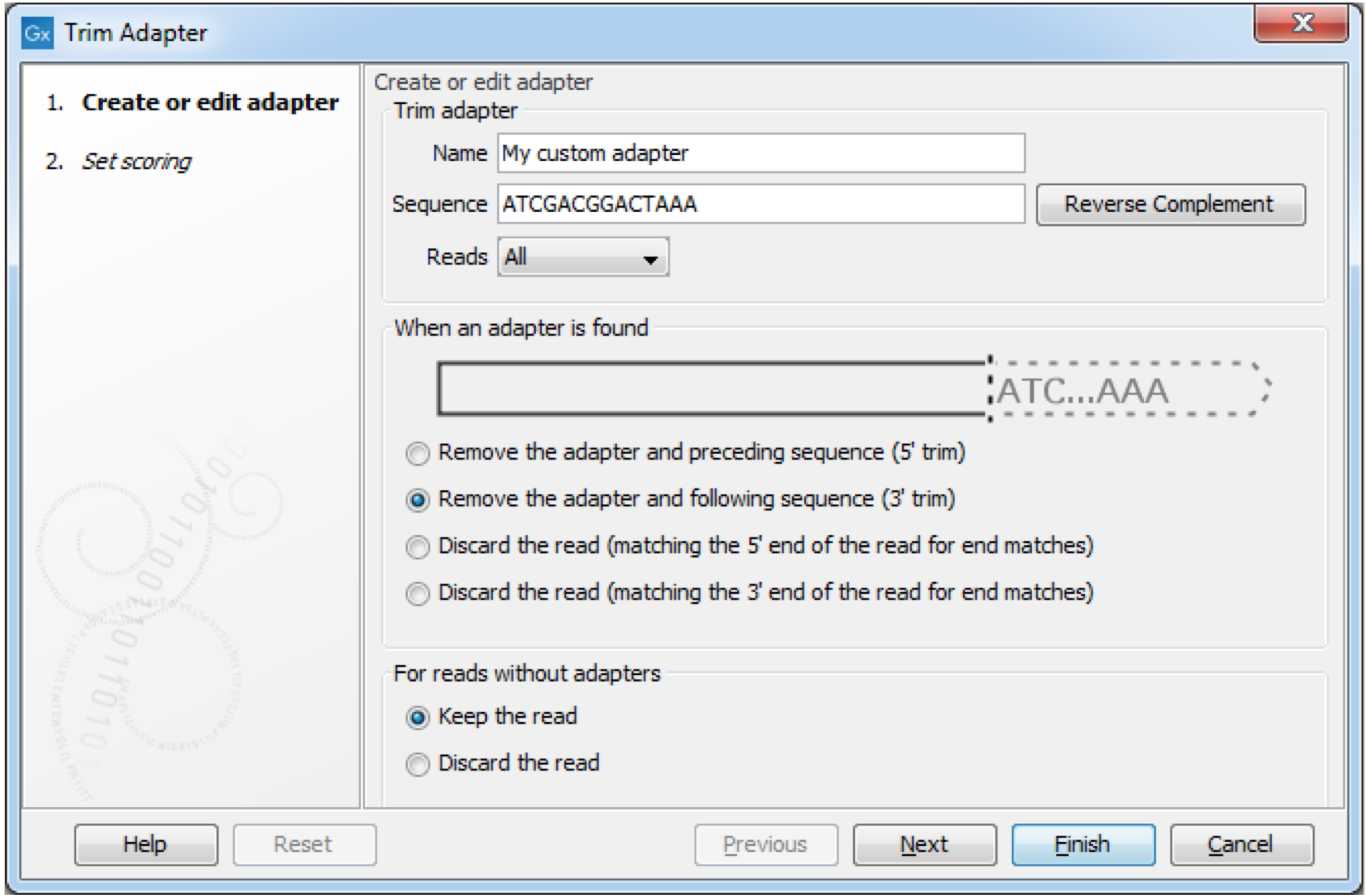
Improved NGS read handling and standard adapter-trimming
It is now easier to import and prepare your NGS reads. We have redesigned the user interface for creating adapter trim lists to improve ease-of-use for defining standard or custom adapters.
- A dynamic graphic now visualizes how the selections during setup will affect the tool’s trim behavior.
- The redesigned interface enables an even wider range of adapter trim-scenarios, but with fewer settings and options for selection.
And many other improvements....
- The speed of the Illumina High-Throughput Sequencing Import has been substantially improved.
- Paired-end reads can now be exported into two FastQ files – one for the first read, and one for the second read. The interleaved format can still be chosen as an export option.
- Updated NCBI Blast executables.
Please see the release notesfor a full list of the many improvements and changes for this release.