
NGS Secondary and Tertiary Analysis
Accelerate patient diagnosis with QCI Interpret AI-enabled platform
The industry’s only automated FASTQ to final report solution for inherited diseases powered by AI-enabled literature searches and state-of-the-art manual curation
Targeted gene panels | Whole exome sequencing | Whole genome sequencing
QCI Interpret for Hereditary Diseases
An end-to-end solution for NGS variant analysis, interpretation and reporting
QCI Interpret for Hereditary Diseases is an end-to-end solution for NGS data analysis, interpretation, and reporting that helps clinical diagnostic labs scale the process of FASTQ to final report. Comprised of two integrated software applications, QCI Interpret for Hereditary alleviates the complexities of regulating in-house bioinformatics pipelines to improve diagnostic yield, reduce turnaround time, and support confident decisions.
QCI Interpret for Hereditary
Cloud-based NGS secondary analysis that is an optional service for labs needing a FASTQ to VCF solution.
Clinical decision support software (tertiary analysis) for labs needing a VCF to final report solution.
Automate FASTQ to final report
1. Upload raw FASTQ sequencing files
• Select pre-configured analysis pipelines in QCI Secondary Analysis (optional service for FASTQ to VCF)
• Generate high-quality variant calls, visualize results, and send VCF files directly to QCI Interpret

2. Upload VCFs and enter patient information
• Select workflow in QCI Interpret
• Upload variant files for a patient sample and enter relevant case details
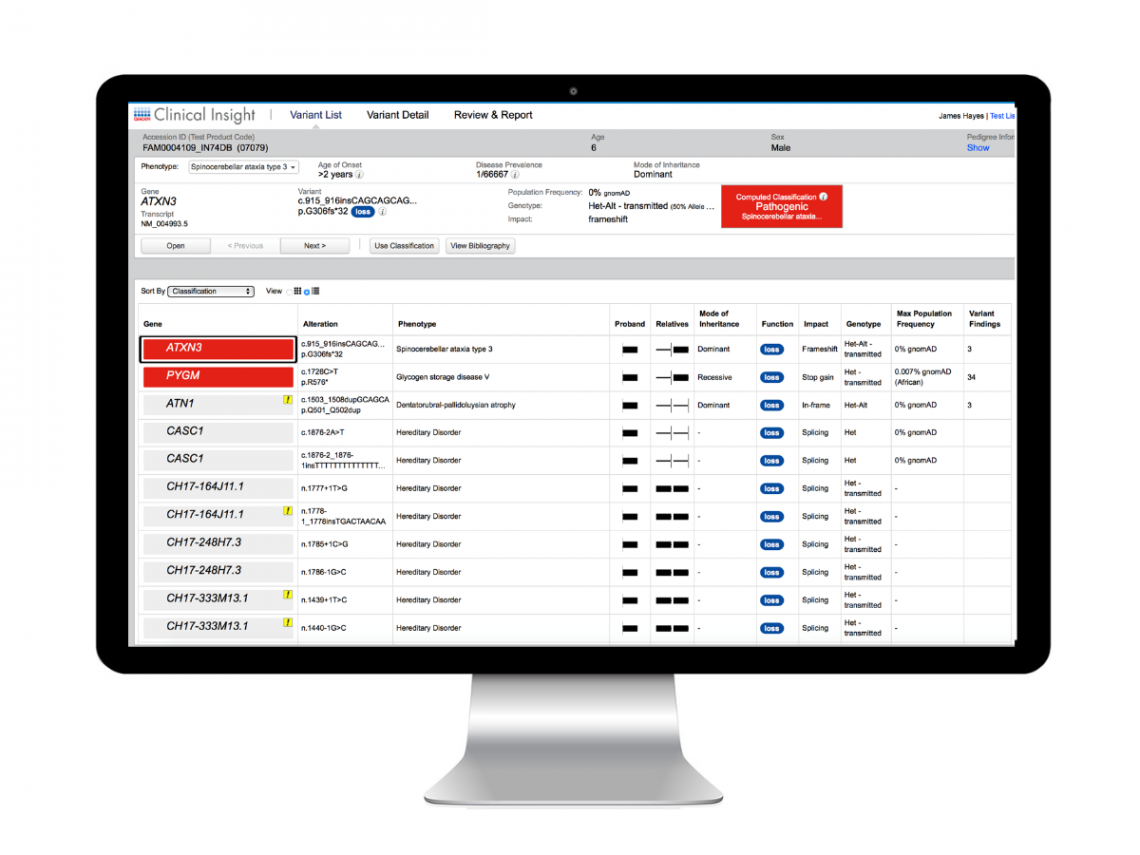
3. Retrieve list of auto-classified variants
- View list of ACMG/AMP classifications for each variant
- Use AI-trained phenotype driven ranking to filter variants by significance

4. Review evidence and set reportability status
- Filter publications to focus exclusively on phenotype-related references
- Evaluate curated data to make final decision on the pathogenicity assessment and reportability status

5. Generate and sign-off on final report
- Generate a final clinical report that is patient-specific and includes clinically relevant variants, interpretations, and references specified throughout the assessment process.
Request a free demo of QCI Interpret for Hereditary
QCI Secondary Analysis
QCI Secondary Analysis is an optional cloud-based service that processes FASTQ files to produce VCF files containing single nucleotide variants (SNV), insertion–deletion mutation (InDel) and structural variant (SV) calls. The module performs quality and adapter trimming, read mapping, deduplication, local realignment, quality control and variant calling and seamlessly connects to QCI Interpret for an integrated and automated FASTQ to report workflow.
- Unrivalled speed -Requires less than 20 minutes to process one WGS sample at 35x coverage.
- Lowest cost per sample –Costs less than $0.40 per WGS sample.
- Accurate –Achieves 99% accuracy for more than 90% of the genome.
QCI Interpret for Hereditary
QCI Interpret for Hereditary is clinical decision support software that combines the unmatched accuracy and consistency of QIAGEN’s proprietary expert (MD/PhD) curation with the superior efficiency of machine curation (AI-powered curation) to enable high-confidence variant interpretation and reporting. Advanced features enable clinical diagnostic labs to rapidly identify pathogenic variants, improve diagnostic yields, and reduce turnaround times. Panel- and sequencer-agnostic, QCI Interpret for Hereditary can be fully customized to accommodate gene panels, exomes, and genomes, with our unrivalled knowledge base providing complete bibliographic coverage of the clinical exome.
Software features
Explore the features of QCI Interpret
With its latest release, QCI Interpret advances its curation approaches with cutting-edge AI content extraction for thousands of rare disease genes to provide complete bibliographic coverage of the clinical exome. Therefore, with this new feature, QCI Interpret customers receive human-certified content for more than 1,000 routinely tested genes that QIAGEN’s curation team monitors daily and AI-derived literature references for the remaining genes in the clinical exome to supplement QIAGEN curated content. In the above screen grab, the green icons indicate human-certified content and the blue icons indicated AI-derived content.
AI-trained phenotype driven ranking tested on a dataset of thousands of solved cases provides superior overall candidate ranking for causative variants in rare diseases. The new variant-ranking approach takes into account multiple variant-specific variables, including the variant’s computed pathogenicity, mode of inheritance, zygosity, patient symptoms, as well as all the manually curated, disease-specific evidence in the QIAGEN Knowledge Base. Greatly enhancing the diagnostic power of exome sequencing, the AI-trained phenotype-driven ranking feature improves the accuracy and efficiency of identifying disease-gene associations.
QCI Interpret leverages advanced AI capabilities that identify all the literature, case reports and clinical trials relevant to your patient’s phenotype with one-click. The software has a new column with “Source” icons (1) with the option to sort (2) and disease “Context” details (when present) to all references (3). The new Show Phenotype-Related References Only filter option narrows down the results to focus exclusively on phenotype-related references (4).
QCI Interpret provides evidence to trigger all 28 criteria of the ACMG/AMP variant interpretation guidelines. Once a VCF file has been uploaded to the software, within seconds, QCI Interpret returns evidence categorized into one of the 28 defined criteria set forth by the ACMG/AMP guidelines and assigns a calculated strength of the evidence. ACMG classifications automatically include case-level information such as inheritance models and relevant findings in associated samples, and if additional information or expertise is available, users can incorporate their data, modify criteria, and store changes for all downstream cases. Users can then view each piece of evidence used in the assessment through clickable hyperlinks that show the full article—not the abstract.
Pathogenicity classifications in QCI Interpret are accompanied by clear visibility into the evidence and criteria supporting the classifications. In addition, you can manually add a rationale for each of the rules triggered if desired and adjust the strength of the final assessment if additional data is available. ‘
QCI Interpret provides the most up-to-date, expertly curated extensive bibliography coverage (published clinical studies, including treatment and prognostic studies, functional studies, review articles, and other types) with multiple lines of evidence linking variants to a disease. Bibliography is categorized by phenotype association for every variant detected and includes all findings that have been manually and AI curated from the published literature as well as findings that have been imported from specific databases. Users can evaluate the curated data to make a final decision on the pathogenicity assessment and reportability status (allow the variant to be displayed on the final clinical report). You can easily add your own criteria for the final variant assessment in the Assessment window
QCI Interpret for Hereditary provides customizable protocols which enable labs to standardize genetic tests and analysis workflows for whole genome sequencing, exome sequencing, and gene panels. Protocols can include combinations of simple or complex filters on any set of annotation entities, can define gene lists or genomic regions, and can be locked down on the user and annotation level. Protocols can also be optimized to automatically render reports and include any user interpretations for given variants. Default protocols are provided for both germline and somatic NGS testing. This allows all users to quickly become proficient in the system and provide a template to optimize.
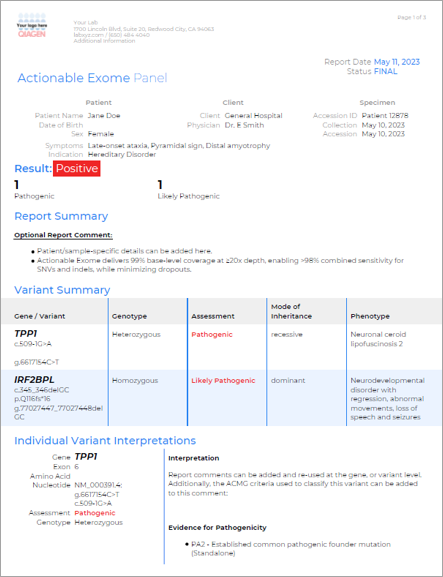
Sample report
QCI Interpret for Hereditary sample report
QCI Interpret provides a comprehensive and flexible reporting system that automatically incorporates significant variants, key findings, annotation sources, and interpretation summaries. Reports can be fully customized to meet your lab’s brand and formatting requirements. This sample report is for an actionable exome panel and shows results with a TPP1and IRF2BPL alteration detected.



Over 3.5 million NGS patient cases interpreted worldwide
Discover why QCI Interpret is one of the most trusted clinical bioinformatics platforms in the world.
Related NGS testing solutions
HGMD Professional
QCI Interpret for Oncology
Product Disclaimer: QCI Interpret is an evidence-based decision support software intended as an aid in the interpretation of variants observed in genomic next-generation sequencing data. The software evaluates genomic variants in the context of published biomedical literature, professional association guidelines, publicly available databases, annotations, drug labels, and clinical-trials. Based on this evaluation, the software proposes a classification and bibliographic references to aid in the interpretation of observed variants. The software is NOT intended as a primary diagnostic tool by physicians or to be used as a substitute for professional healthcare advice. Each laboratory is responsible for ensuring compliance with applicable international, national, and local clinical laboratory regulations and other specific accredidations requirements.