
Variants covering the exact length of a target region are not called and variants are not called for target regions of 1bp
Issue description
When calling variants with the Basic Variant Detection, Low Frequency Variant Detection, or Fixed Ploidy Variant Detection tool and using the “Restrict calling to target regions” option, the following problems exist:
- Any variant that is exactly the length of the target region will not be called. For example, if a target region is made that is 1bp long, with the intention of detecting a known SNV at that position, that SNV will not be detected. Similarly, any target region defined such that it covers exactly the length of a variant will be affected by this problem.
- No variants will be called for target regions of 1 bp regardless of the length of the variant.
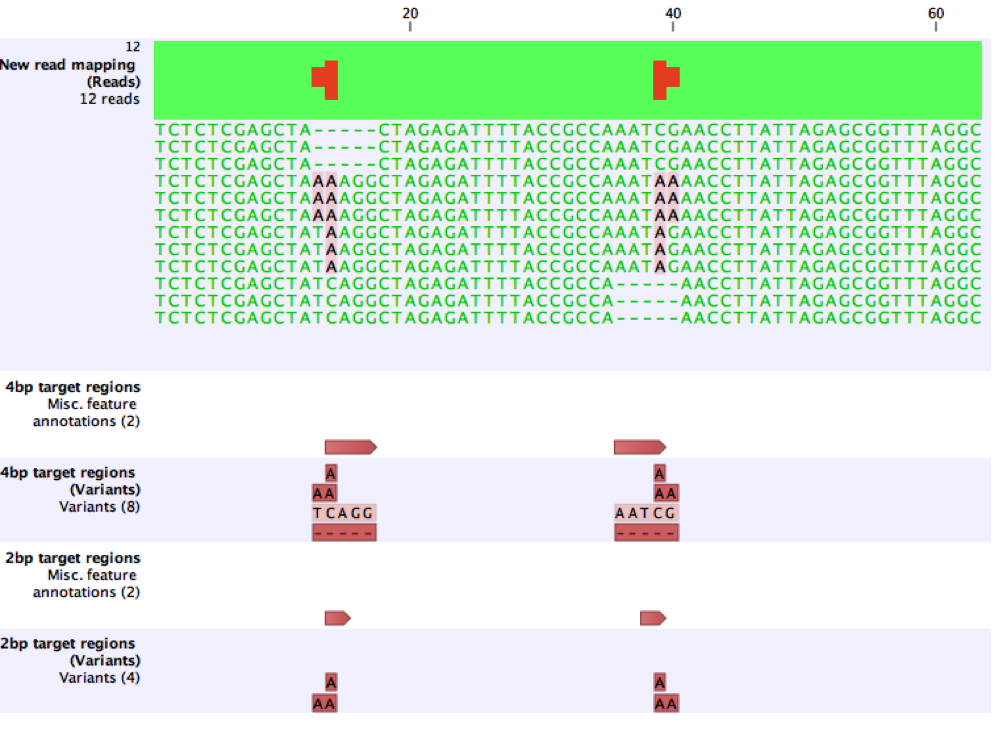
The effect of these issues are illustrated using a small, synthetic example. Here, 4bp target regions are provided, and six variants are called as expected. After reducing the target regions to 2bp, the 2bp MNVs, AG (left) and CA (right) are not called because they exactly match the coordinates of the target regions. When the target regions are further reduced to 1bp no variants are called.
Affected software
- CLC Genomics Workbench 6.5 to CLC Genomics Workbench 12.0.1
- CLC Genomics Server 5.5 to CLC Genomics Server 11.0.1
- All versions of Biomedical Genomics Workbench
This issue was fixed in CLC Genomics Workbench 12.0.2 and CLC Genomics Server 11.0.2.