For low- to mid-throughput labs
Complete NGS workflows for Complete Genomics MGI G99 benchtop sequencer
Does your lab need an out-of-the-box, simple-to-use, sample-to-report NGS workflow?
For mid-throughput labs who are interested in using the Complete Genomics (MGI) DNBSEQ-G99 benchtop sequencer, QIAGEN provides sample-to-report NGS workflows with targeted DNA panels and out-of-the-box downstream data analysis software solutions.
Disclaimer: QIAGEN does not have a partnership with MGI; rather this workflow is for demo purposes only.

Build your workflow with four NGS steps
Step 1: Library Preparation
Step 2: Sequencing
Step 3: Data Analysis
Step 4: Interpretation and Reporting
Step 1: Library Preparation
QIAseq Targeted DNA Pro Panels
Learn more about QIAseq's state-of-the-art enrichment technology
QIAseq Targeted DNA Pro Panels set a new standard for NGS genotyping, providing improved ease-of-use, efficiency and expanded variant analysis. Each panel is a one-box, NGS platform-agnostic solution that contains all the necessary components to construct libraries from enriched genomic targets. This highly optimized DNA target enrichment technology offers an automation-friendly solution that simultaneously facilitates ultrasensitive variant detection using integrated unique molecular indices (UMIs).
- Rapid turnaround time – Streamlined library preparation workflows take less than 6 hours with minimal hands-on time.
- Ultrasensitive variant detection – Uses integrated unique molecular indices UMIs to detect genetic variations, including somatic and germline mutations, single nucleotide polymorphisms, copy number variation and small insertions/deletions.
- Cost efficient – Maximized read utilization results in up to 50% reduction in sequencing costs
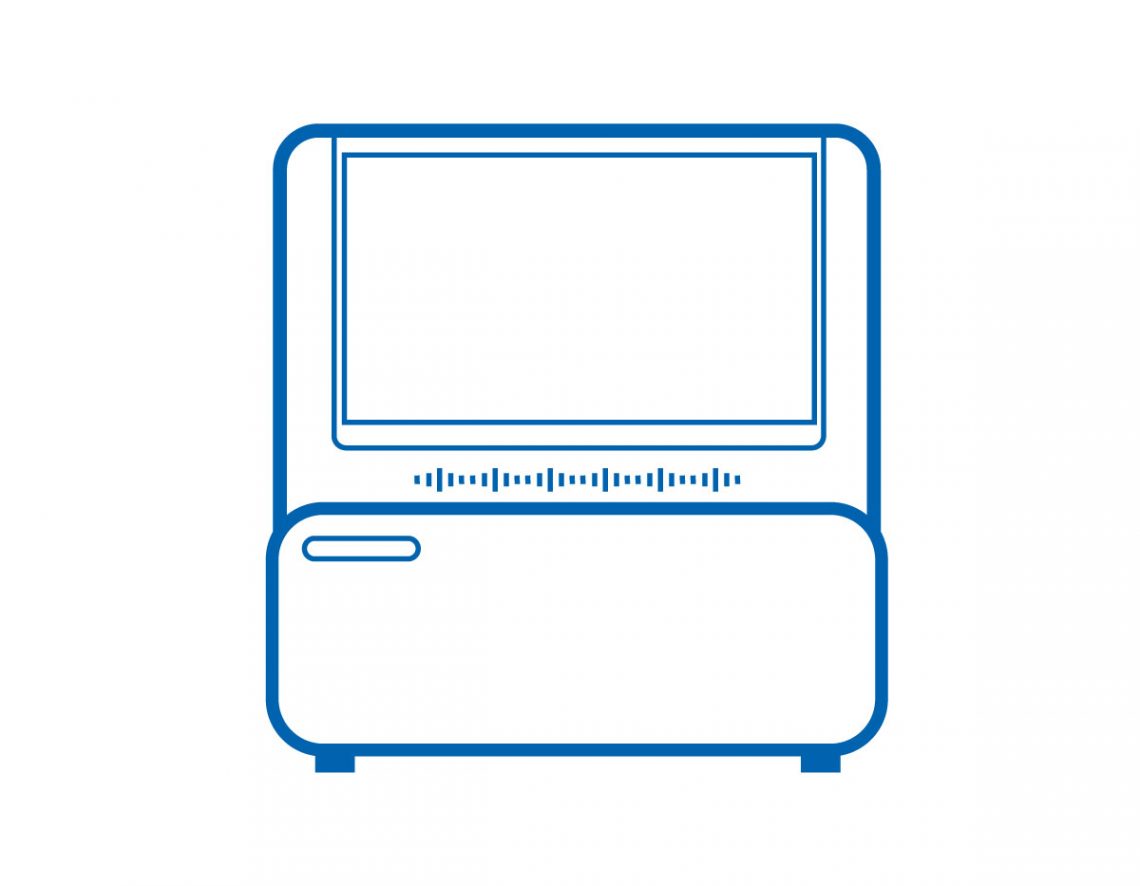
Step 2: Sequencing
Complete Genomics (MGI) DNBSEQ-G99 benchtop sequencer
As one of the sequencers with the fastest speed among those with the same throughput in the world, DNBSEQ-G99 is especially suitable for targeted gene sequencing and small genome sequencing, delivering rapid and high-quality data output. Based on MGI’s DNBSEQTM core technology, DNBSEQ-G99 for adopts triangular matrix signal spots on sequencing flow cell, reaching higher density of data output, with an overall throughput of 8-48 Gb per run. The instrument also provides rapid sequencing, enabling users to go from loading to FASTQ within 12 hours.
- Ultra-fast – Requires only 12 hours for whole PE150 sequencing (from loading to FASTQ).
- Flexible data output – Dual flow cell design enables users to achieve flexible data output on one sequencer* while supporting multiple sequencing modes.
- Simple operation – Provides automatic analysis and result output, clean and concise visual design, and localized data storage and calculation.
Step 3: Data Analysis
QCI Secondary Analysis with LightSpeed
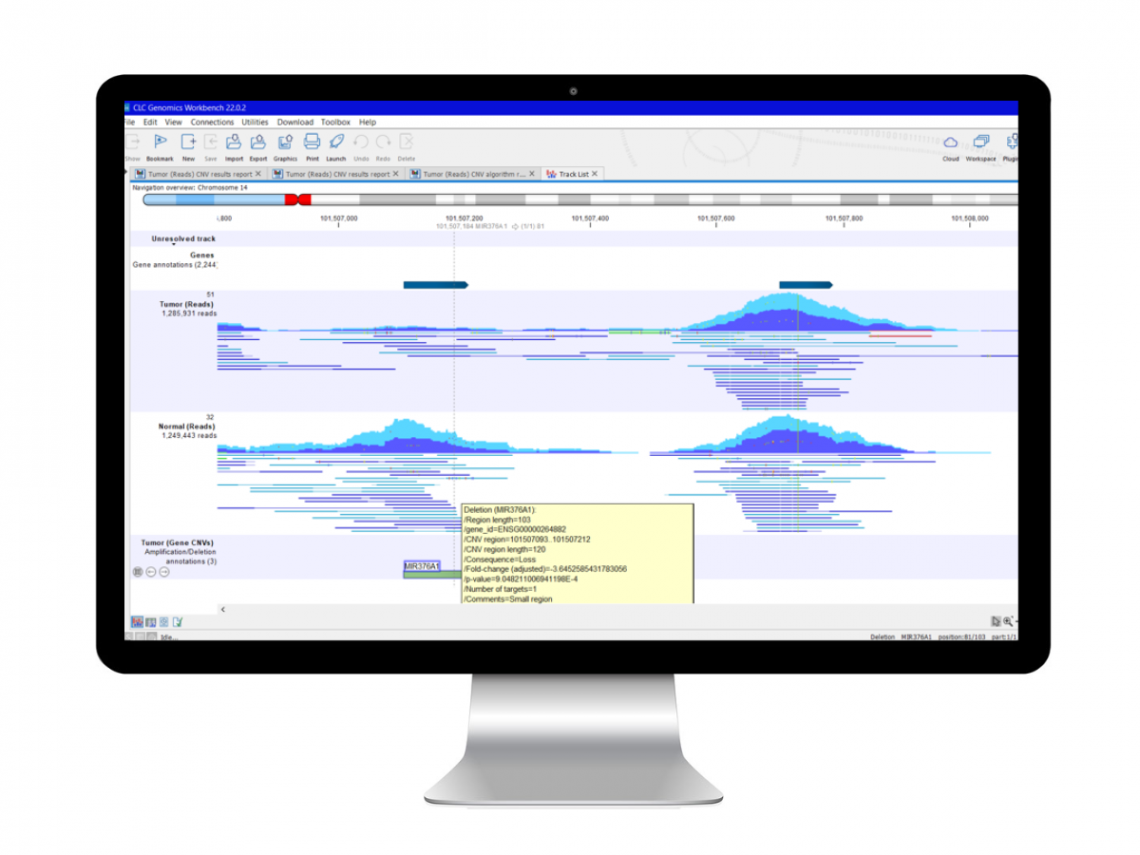
LightSpeed computes 30% faster than Illumina® DRAGEN®
LightSpeed is a new module for QIAGEN CLC Genomics Workbench Premium that empowers laboratories to perform NGS secondary analysis with high accuracy at unprecedented runtimes. FASTQ files produced from the AVITI™ System are uploaded to LightSpeed. Within minutes (or less), LightSpeed produces VCF files containing single nucleotide variants (SNV), insertion–deletion mutation (InDel) and structural variant (SV) calls. The module is deployable using local computers or Amazon Web Services (AWS®) cloud and performs quality and adapter trimming, read mapping, deduplication, local realignment, quality control and variant calling.
- Agnostic – Analysis workflows are custom-tailored to panels from any vendor.
- Ultra-rapid – Requires less than 20 minutes to process one WGS sample at 35x coverage.
- Cost-efficient – Significantly reduces computing costs, requiring less than $0.40 per WGS sample.
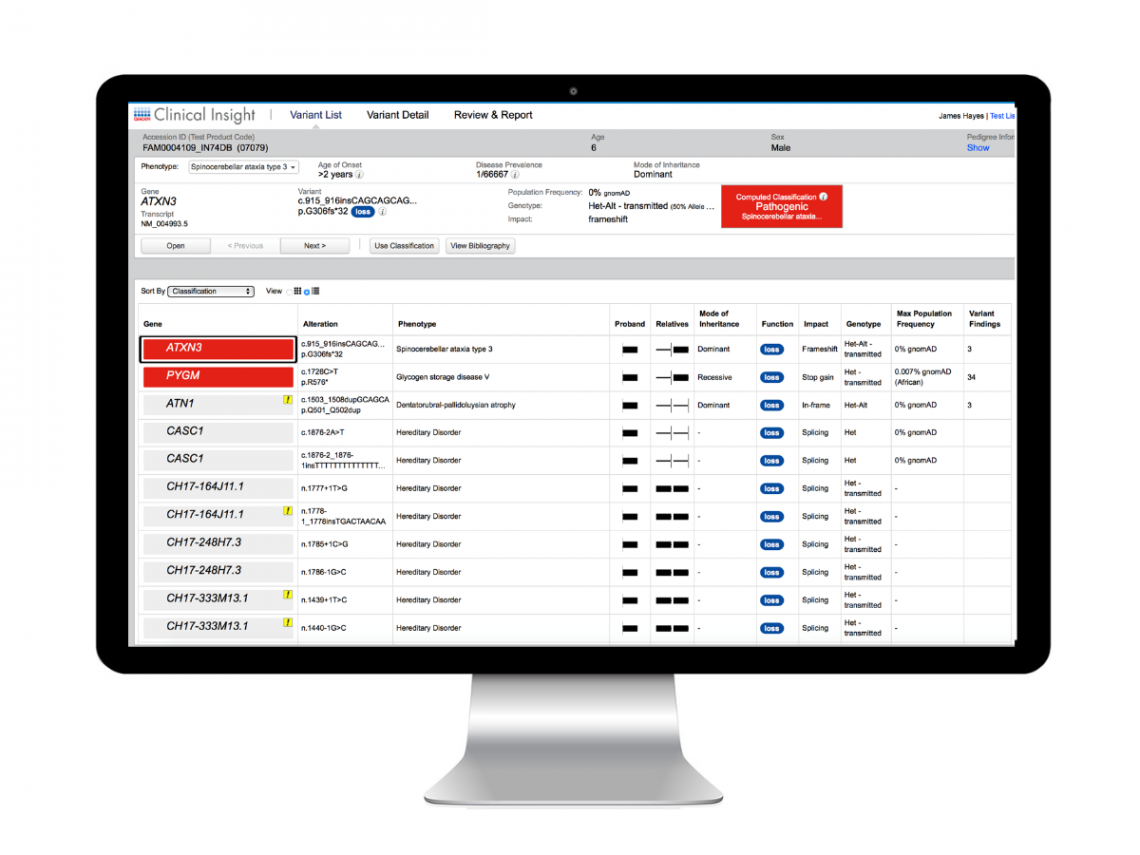
Step 4: Interpretation and Reporting
QCI Interpret
QCI Interpret is an out-of-the-box, fully customizable software solution that enables NGS variant interpretation and reporting for oncology and hereditary applications. VCF files from QCI Secondary Analysis are directly uploaded to QCI Interpret. Users then select a preconfigured workflow, and QCI Interpret automatically filters variants and computes classifications according to ACMG and AMP/ASCO/CAP professional guidelines.
Unlike other tertiary analysis solutions, QCI Interpret uses augmented molecular intelligence (the combination of human and machine curation) to ensure the evidence being considered is accurate, consistent and up-to-date.Users can then review the literature and criteria supporting each variant classification, determine reportability status, and confidently generate a final, patient-specific report with trusted diagnostic, prognostic and therapeutic information.
View a sample report here.
Unrivalled content
QCI Interpret’s knowledge base encompasses >40 databases maintained by machine and human curators who add >46,000 new findings each week.
Cost-saving efficiency
By automating variant annotation, classification, and curation, as well as the report building process, QCI Interpret enables significant time and cost savings.
Confident reporting
QCI Interpret ensures you deliver patient-specific reports backed by the latest scientific and clinical evidence and therapeutic options (on and off label).
Resources
Application Note
Webinar
Sample Report
Contact us
Let us help you optimize your NGS analysis pipeline. Our services team is here to answer your questions and help you get from sample to final report in less time, for less money.