Sample to Insight for Inherited Disorders
Break speed and cost barriers
in genetic testing
Does your lab need a fast, cost-effective, ultra-precise workflow for germline NGS testing?
Next-generation sequencing (NGS) has transformed the field of genetic testing, enabling rapid and cost-effective identification of genetic variants associated with inherited diseases. QIAGEN’s new groundbreaking Sample to Insight solutions for inherited diseases combine human exome and human hybrid-capture panels, the fastest and cheapest secondary analysis in the market, and trusted variant interpretation and reporting software powered by augmented molecular intelligence.
Sample to Insight for Inherited Disorders:
On-demand webinar
Panels
QIAseq Human Exome and xHYB Human Hybrid Capture Panels
The QIAseq portfolio provides flexible options for hybrid-capture based human exome sequencing assays, including a 34 Mb exome panel, and curated content that targets variants in over 10,000 genes from the Human Gene Mutation Database (HGMD). And, optimized probe design provides efficient coverage of even the most challenging genomic regions, such as high GC-content regions.
- Never miss a target – Panels for covering HGMD-curated whole genes and protein coding sequences (CDS)
- Complete, uniform coverage – delivers 99% base-level coverage at ≥20x depth, enabling >98% combined sensitivity for SNVs and indels, while minimizing dropouts
- Single day, automation-friendly workflow – Single-day workflow with hybrid capture flexibility ranging from 30 minutes to overnight
- Reduce costs – Maximized read utilization results in up to 50% reduction in sequencing costs
Delivers excellent coverage uniformity regardless of the target GC composition for sensitive detection of pathogenic variants, enabling time and cost efficiencies critical to scalability.
Panel size: 33.9 Mb
Genes: 10,000+
Targets all (100%) disease-causing and likely disease-causing variants listed in HGMD (>300,000 variants), including variants in regulatory regions and splice sites, as well as deep intronic variants.
Panel size: 12.3 Mb
Genes: 10,000+
A hybrid-capture based solution for comprehensive carrier screening that adheres to the latest recommendations for screening autosomal recessive and X-linked conditions given by ACMG.
Panel size: 1.5 Mb
Genes: 448
Provides a spike-in to cover the full mitochondrial genome and can be paired with exome or other panels.
Panel size: 16.6 kb
Genes: Full mitochondrial genome
NGS Secondary Analysis
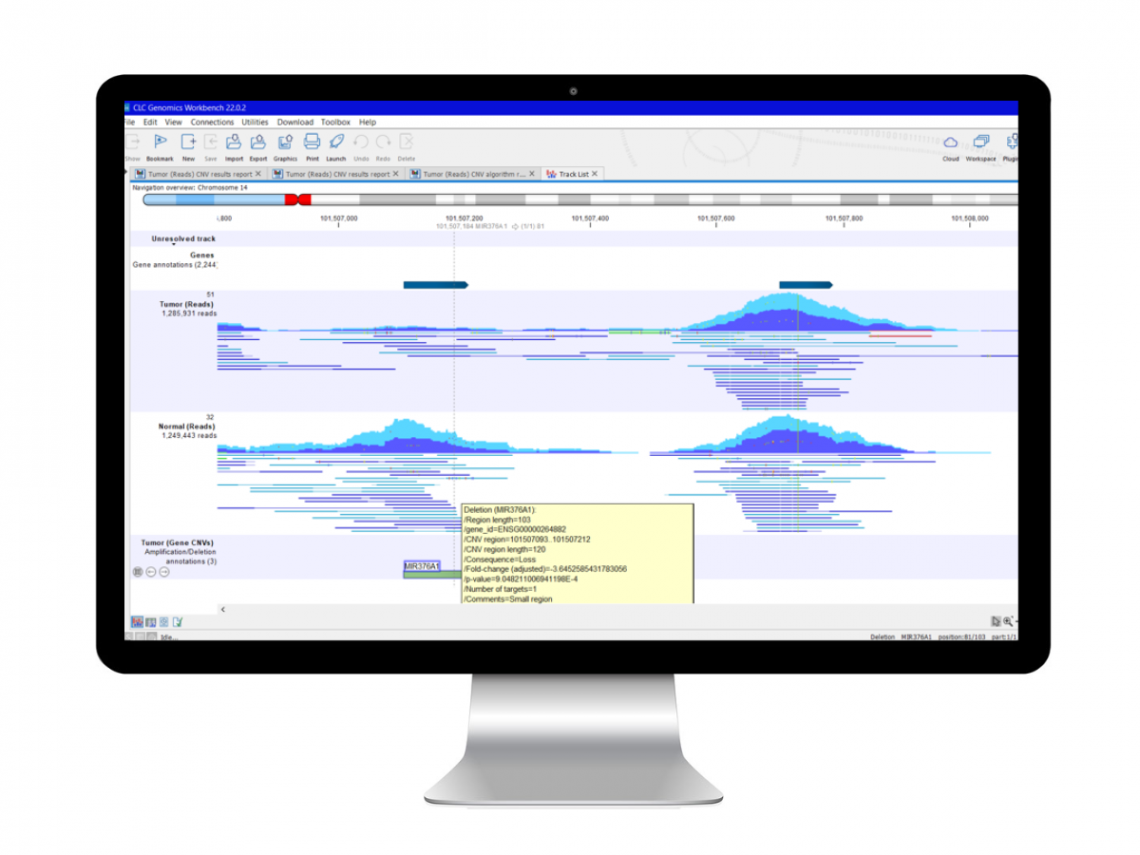
QCI Secondary Analysis with LightSpeed
LightSpeed is a new module for QIAGEN CLC Genomics Workbench Premium that empowers laboratories to perform NGS secondary analysis with high accuracy at unprecedented runtimes.
LightSpeed processes FASTQ files to produce VCF files containing single nucleotide variants (SNV), insertion–deletion mutation (InDel) and structural variant (SV) calls. The module is deployable using local computers or Amazon Web Services (AWS®) cloud and performs quality and adapter trimming, read mapping, deduplication, local realignment, quality control and variant calling.
Variant Interpretation and Reporting
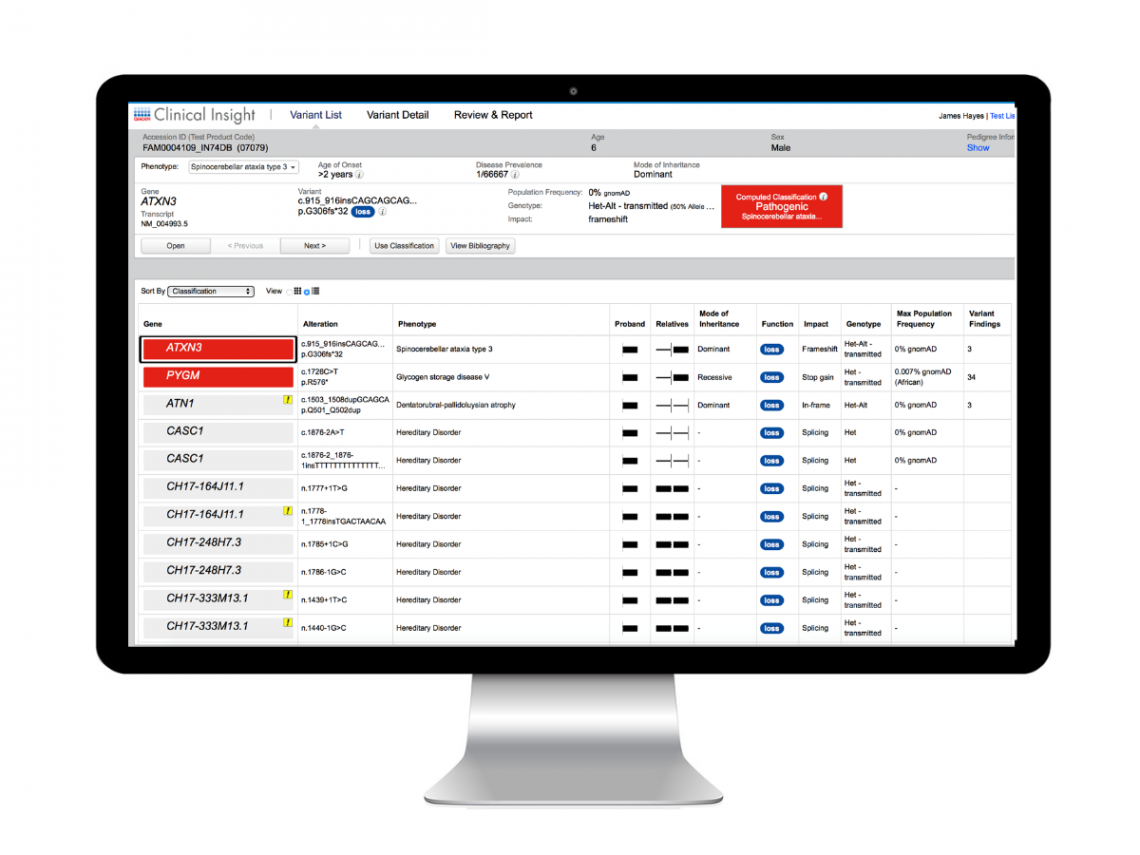
QCI Interpret for Hereditary
QCI Interpret is NGS variant interpretation and reporting software powered by augmented molecular intelligence that helps labs not only make faster decisions—but the right decisions.
Connected to the exclusive QIAGEN Knowledge Base, the industry’s most comprehensive, manually curated resource updated weekly, QCI Interpret delivers variant-specific, scientific evidence in context of phenotype or diagnosis. Interactive filters prioritize variants and proprietary algorithms transparently compute ACMG/AMP variant classifications, enabling users to generate evidence-based reports with efficiency, confidence, and reproducibility.
Resources
Contact us
Let us help you optimize your NGS analysis pipeline. Our services team is here to answer your questions and help you get from sample to final report in less time, for less money.

Disclaimers:
The QIAseq Human Exome and xHYB Human Hybrid Capture Panels are intended for molecular biology applications. This product is not intended for the diagnosis, prevention or treatment of disease.
LightSpeed is software module to support NGS secondary analysis. The software is NOT intended as a primary diagnostic tool by physicians or to be used as a substitute for professional healthcare advice. Each laboratory is responsible for ensuring compliance with applicable international, national, and local clinical laboratory regulations and other specific accreditations requirements.
QCI Interpret is an evidence-based decision support software intended as an aid in the interpretation of variants observed in genomic next-generation sequencing data. The software evaluates genomic variants in the context of published biomedical literature, professional association guidelines, publicly available databases, annotations, drug labels, and clinical trials. Based on this evaluation, the software proposes a classification and bibliographic references to aid in the interpretation of observed variants. The software is NOT intended as a primary diagnostic tool by physicians or to be used as a substitute for professional healthcare advice. Each laboratory is responsible for ensuring compliance with applicable international, national, and local clinical laboratory regulations and other specific accreditations requirements.