
Variant Analysis 2015 fall release notes
Whole genome prefilter limit increased from 200 to 300 samples
To improve performance, users have the option to pre-filter their whole genome data to only exonic regions. When an analysis exceeds a certain volume of whole genome samples, pre-filtering is mandatory. With the fall release, the upper limit that imposes mandatory prefiltering has been lifted from 200 to 300 whole genome samples. Now users performing analysis up to 300 whole genomes are not required to pre-filter. Users creating analysis with greater than 300 whole genomes must use pre-filter or contact Customer Support to create analysis without pre-fltering.
Private Control Libraries
Private Control Libraries (PCL) will allow users to compute variant frequency from a select set of samples and allow users to filter on those frequencies as well as compare case sample(s) vs all the samples in the PCL. The PCL will allow analysis with large volume of control samples to compute faster and enable much larger analysis sizes (up to 2000 whole genomes). One can also compare case(s) vs control samples within the PCL using the Genetic Analysis and Statistical Analysis filters.
The IVA application will feature a new tab “My Control Libraries” where you can see your collection of Private Control Libraries. Additionally there will be a new “New Library” button in the My Samples view to build new libraries.
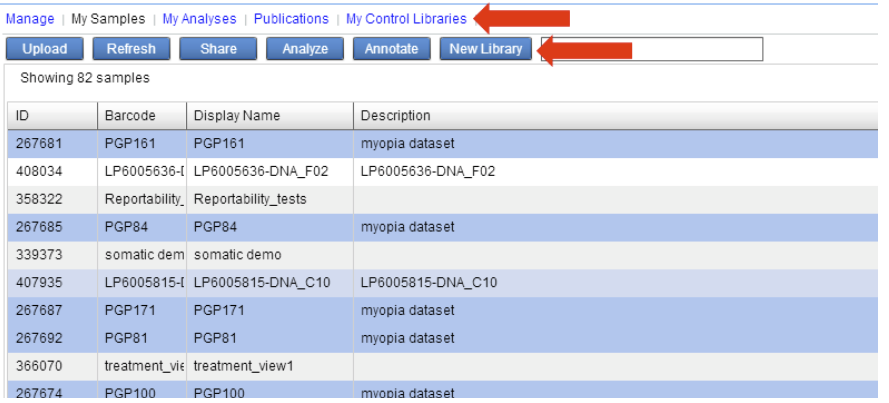
When creating a new private control library, users can build a library by selecting and dragging the samples from the left pane to the right pane of the Create Control Library window.
After clicking the “Create” button, the library is processed offline and the user will be notified by email when the PCL is built. When the library is built, users can access and review the library using the My Control Libraries tab which will list all existing PCLs. From here one can edit the library’s meta data or delete the library.
Updated build of Allele Frequency Community
The new build of the Allele Frequency Community (AFC) is based off a collection of 120,000 consented exomes and genomes. Of the 120,000 consented samples, roughly 12,000 are whole genomes and the rest are exomes.
Improved (beta) integration with IPATM
With the fall release, we have improved the integration between IVA and IPA. Now when you click on the Export to IPA button, IVA will export the list of gene IDs, the ACMG assessments, and the gain or loss of function information. This features is in beta and would have limited support for the fall release.